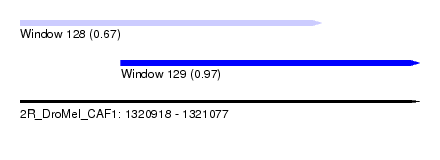
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,320,918 – 1,321,077 |
| Length | 159 |
| Max. P | 0.970403 |

| Location | 1,320,918 – 1,321,038 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.87 |
| Mean single sequence MFE | -47.04 |
| Consensus MFE | -33.83 |
| Energy contribution | -33.67 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.672018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
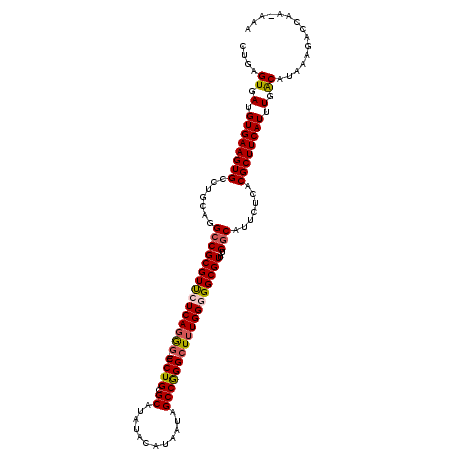
>2R_DroMel_CAF1 1320918 120 + 20766785 ACGCUCGCCCCCCUCCCUACCCCUUGACGAGGCGCCCCUGUUGAGUGAUGUGAAGUGCCUGCAGGCCGCGUUCUCAGGGCCGCCUGCGCAUAUACAUAAUAGCCAGGCUUUGGGGGCGUU .....(((((((.........((((((.((.(((..(((((.(.(((........)))).))))).))).)).))))))..(((((.((.(((.....))))))))))...))))))).. ( -47.20) >DroSec_CAF1 39454 106 + 1 CCGCUCGCCCCCUUC-----------CCGGGGCGCCCCUGCUGAGUGAUGUGAAGUGCCUGCAGGCCGCGUUCUCAGGG---CCUGCGCAUAUACAUAAUAGCCGGGCUGUGGGGGCGUU .....(((((((..(-----------(((((....))).((((....(((((..((((..(((((((.(.......)))---))))))))).)))))..)))).)))....))))))).. ( -50.80) >DroSim_CAF1 18558 106 + 1 CCGCUCGCUCUCUUC-----------ACGGGGCGCUCCUGCUGAGUGAUGUGAAGUGCCUGCAGGCCGCGUUCUCAGGG---CCUGCGCAUAUACAUAAUAGCCGGGCUUUGGGGGCGUU ..((..((.(.((((-----------(((...(((((.....))))).))))))).)...))..)).((((((((((((---((((.((.(((.....))))))))))))))))))))). ( -47.70) >DroEre_CAF1 20952 97 + 1 CCGCUCGCCCCUUUU--------------GGGCGUGCCUGCUGAGUGAUGUGAAGUGCCUGCAGG-CGCGUCCUCAGAU---CCUGCGCAUA----UAAUAGCCGGGCUUUGG-GGCGUU .....((((((....--------------((((((((((((.(.(((........)))).)))))-)))))))..(((.---((((.((...----.....)))))).)))))-)))).. ( -45.00) >DroYak_CAF1 6953 100 + 1 CCGCUCGCCCCCUUU------------CGGGGCGCCCCUGUUGAGUGAUGUGAAGUGCCUGCAGGCCGCGUUUUCAGGG---CCUGCGCAUA----UAAUAGCCAGGCUUUGG-GGCGUU ......(((((....------------.)))))(((((....(((.((((((....(((....))))))))))))((((---((((.((...----.....))))))))))))-)))... ( -44.50) >consensus CCGCUCGCCCCCUUC____________CGGGGCGCCCCUGCUGAGUGAUGUGAAGUGCCUGCAGGCCGCGUUCUCAGGG___CCUGCGCAUAUACAUAAUAGCCGGGCUUUGGGGGCGUU ......(((((.................)))))((((((.(((((.((((((....(((....)))))))))))))).....((((.((............))))))....))))))... (-33.83 = -33.67 + -0.16)

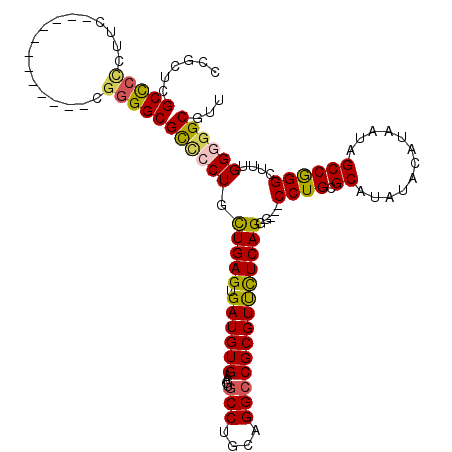
| Location | 1,320,958 – 1,321,077 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.63 |
| Mean single sequence MFE | -40.95 |
| Consensus MFE | -33.48 |
| Energy contribution | -33.68 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1320958 119 + 20766785 UUGAGUGAUGUGAAGUGCCUGCAGGCCGCGUUCUCAGGGCCGCCUGCGCAUAUACAUAAUAGCCAGGCUUUGGGGGCGUUAUGGCAUUCUCACGCUUCAUUUGGCAUAAAGACCAA-AAA .((((.(((((.((((((((.((((((.(.......)))))(((((.((.(((.....))))))))))..)).))))))).).))))))))).......(((((........))))-).. ( -39.40) >DroSec_CAF1 39483 116 + 1 CUGAGUGAUGUGAAGUGCCUGCAGGCCGCGUUCUCAGGG---CCUGCGCAUAUACAUAAUAGCCGGGCUGUGGGGGCGUUAUGGCAUUCUCACGCUUCAUUUGACAUAAAGACCAA-AAA ....((.(.((((((((.......((((((((((((.((---((((.((.(((.....))))))))))).)))))))))...))).......)))))))).).))...........-... ( -43.14) >DroSim_CAF1 18587 117 + 1 CUGAGUGAUGUGAAGUGCCUGCAGGCCGCGUUCUCAGGG---CCUGCGCAUAUACAUAAUAGCCGGGCUUUGGGGGCGUUAUGGCAUUCUCACGCUUCAUUUGGCAUAAAGACCAAUUAA (..((((((((((((((((........((((((((((((---((((.((.(((.....)))))))))))))))))))))...)))))).)))))..)))))..)................ ( -44.50) >DroEre_CAF1 20978 110 + 1 CUGAGUGAUGUGAAGUGCCUGCAGG-CGCGUCCUCAGAU---CCUGCGCAUA----UAAUAGCCGGGCUUUGG-GGCGUUAUGGCAUUCGCACGCUUCAUUUGACAUAAAGACCAA-AAA ..(((((.(((((((((((....))-)))((((.((((.---((((.((...----.....)))))).)))))-))).........)))))))))))...................-... ( -36.70) >DroYak_CAF1 6981 111 + 1 UUGAGUGAUGUGAAGUGCCUGCAGGCCGCGUUUUCAGGG---CCUGCGCAUA----UAAUAGCCAGGCUUUGG-GGCGUUAUGGCAUUCGCACGCUUCAUUUGACAUAAAGACCAA-AAA ....((.(.((((((((..(((..((((((.(..(((((---((((.((...----.....))))))))))).-.))))...)))....))))))))))).).))...........-... ( -41.00) >consensus CUGAGUGAUGUGAAGUGCCUGCAGGCCGCGUUCUCAGGG___CCUGCGCAUAUACAUAAUAGCCGGGCUUUGGGGGCGUUAUGGCAUUCUCACGCUUCAUUUGACAUAAAGACCAA_AAA ....((.(.((((((((.......(((((((((((((((...((((.((............))))))))))))))))))...))).......)))))))).).))............... (-33.48 = -33.68 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:25:25 2006