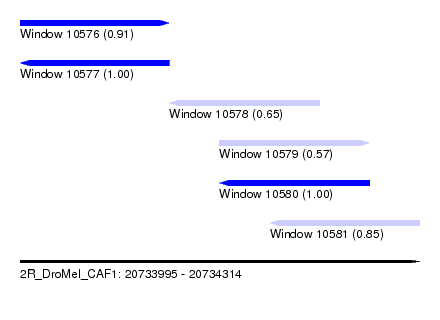
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,733,995 – 20,734,314 |
| Length | 319 |
| Max. P | 0.999725 |

| Location | 20,733,995 – 20,734,114 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.15 |
| Mean single sequence MFE | -34.76 |
| Consensus MFE | -33.56 |
| Energy contribution | -32.80 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20733995 119 + 20766785 ACUCAACUCAGCGAAGUACCCAGUUUUUACUUUACCUGCU-CAGGCUUGGGGAUCGUUUUUCGCUUUCGCCGUGAUGAUUUCCCUUCCUGUCUGCACCCUUAUUUGCGUGCGUGUGAGCG ..........(..(((((.........)))))..)..(((-((.((..((..((((((....((....))...))))))..))..........((((.(......).)))))).))))). ( -30.90) >DroSec_CAF1 167524 119 + 1 ACUCAACUCAGCGAAGUACCCAGUUUUUACUUUACCUGCU-CAGGCUUGGGGAUCGUUUUUCGCUUUCGCCGUGAUGAUUUCCCUUCCUGUUUGCACGCUUAUUUGCGUGCGUGUGAGCG ..........(..(((((.........)))))..)..(((-((.((..((..((((((....((....))...))))))..))..........((((((......)))))))).))))). ( -37.20) >DroSim_CAF1 172942 119 + 1 ACUCAACUCAGCGAAGUACCCAGUUUUUACUUUACCUGCU-UAGGCUUGGGGAUCGUUUUUCGCUUUCGCCGUGAUGAUUUCCCUUCCUGUUUGCACGCUUAUUUGCGUGCGUGUGAGCG ..........(..(((((.........)))))..)..(((-((.((..((..((((((....((....))...))))))..))..........((((((......)))))))).))))). ( -35.00) >DroEre_CAF1 208440 120 + 1 ACUCAACUCAGCGAAGUACCCAGUUUAUACUUUACCUGCUUCGGGCUUGGGGAUCGUUUUUCGCUUUCGCCGUGAUGAUUUCCCUUCCCGUUUGUACGCGUAUUUGCGUGCGUGUGAGCG ......................((((((((...........((((...((..((((((....((....))...))))))..))...))))...(((((((....))))))))))))))). ( -37.50) >DroYak_CAF1 174195 119 + 1 ACUCAACUCAGAGAAGUACCCA-UUUAUACUUUACCUGCUUCGGGCUUGGGGAUCGUUUUUCGCUUUCGCCGUGAUGGUUUCCCUUCCUGUUUGUACGCGUAUUUGCGUGCGUGUGAGCG .((((((.(((.((((((....-....))))))..)))...((((...((..((((((....((....))...))))))..))...))))...(((((((....))))))))).)))).. ( -33.20) >consensus ACUCAACUCAGCGAAGUACCCAGUUUUUACUUUACCUGCU_CAGGCUUGGGGAUCGUUUUUCGCUUUCGCCGUGAUGAUUUCCCUUCCUGUUUGCACGCUUAUUUGCGUGCGUGUGAGCG .((((((.(((.((((((.........))))))..)))...((((...((..((((((....((....))...))))))..))...))))...((((((......)))))))).)))).. (-33.56 = -32.80 + -0.76)

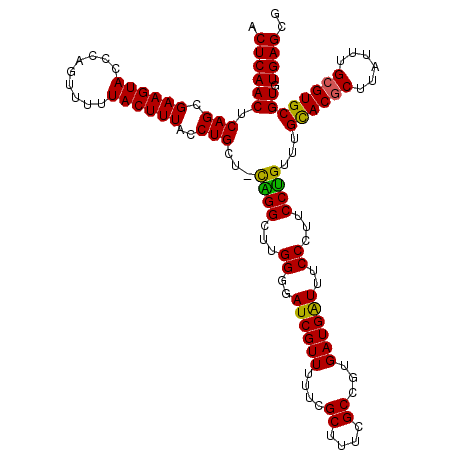
| Location | 20,733,995 – 20,734,114 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.15 |
| Mean single sequence MFE | -35.61 |
| Consensus MFE | -31.10 |
| Energy contribution | -32.14 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.998356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20733995 119 - 20766785 CGCUCACACGCACGCAAAUAAGGGUGCAGACAGGAAGGGAAAUCAUCACGGCGAAAGCGAAAAACGAUCCCCAAGCCUG-AGCAGGUAAAGUAAAAACUGGGUACUUCGCUGAGUUGAGU .(((((.((((((.(......).))))...((((..((((..........((....)).........))))....))))-((((((((.(((....)))...))))).)))..))))))) ( -35.31) >DroSec_CAF1 167524 119 - 1 CGCUCACACGCACGCAAAUAAGCGUGCAAACAGGAAGGGAAAUCAUCACGGCGAAAGCGAAAAACGAUCCCCAAGCCUG-AGCAGGUAAAGUAAAAACUGGGUACUUCGCUGAGUUGAGU .(((((.((((((((......))))))...((((..((((..........((....)).........))))....))))-((((((((.(((....)))...))))).)))..))))))) ( -39.41) >DroSim_CAF1 172942 119 - 1 CGCUCACACGCACGCAAAUAAGCGUGCAAACAGGAAGGGAAAUCAUCACGGCGAAAGCGAAAAACGAUCCCCAAGCCUA-AGCAGGUAAAGUAAAAACUGGGUACUUCGCUGAGUUGAGU .(((((.((((((((......))))))...(((((((.............((....))...........(((..((((.-...))))..(((....))))))..)))).))).))))))) ( -37.10) >DroEre_CAF1 208440 120 - 1 CGCUCACACGCACGCAAAUACGCGUACAAACGGGAAGGGAAAUCAUCACGGCGAAAGCGAAAAACGAUCCCCAAGCCCGAAGCAGGUAAAGUAUAAACUGGGUACUUCGCUGAGUUGAGU .(((((.((((((((......)))).....((((..((((..........((....)).........))))....))))..)).((..((((((.......))))))..))..))))))) ( -35.81) >DroYak_CAF1 174195 119 - 1 CGCUCACACGCACGCAAAUACGCGUACAAACAGGAAGGGAAACCAUCACGGCGAAAGCGAAAAACGAUCCCCAAGCCCGAAGCAGGUAAAGUAUAAA-UGGGUACUUCUCUGAGUUGAGU .(((((.((((((((......)))).......(((.((....))......((....)).........)))....))......((((..((((((...-...)))))).)))).))))))) ( -30.40) >consensus CGCUCACACGCACGCAAAUAAGCGUGCAAACAGGAAGGGAAAUCAUCACGGCGAAAGCGAAAAACGAUCCCCAAGCCUG_AGCAGGUAAAGUAAAAACUGGGUACUUCGCUGAGUUGAGU .(((((.((((((((......))))))...(((((((.............((....))...........(((..((((.....))))..(((....))))))..)))).))).))))))) (-31.10 = -32.14 + 1.04)

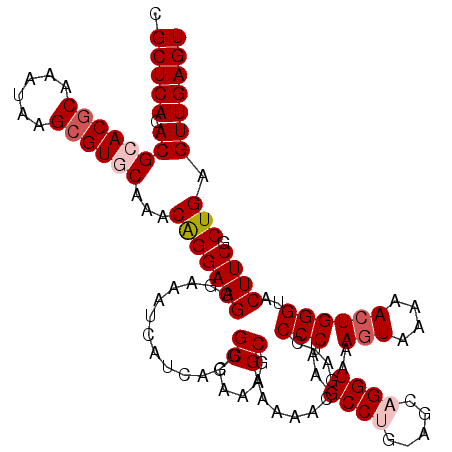
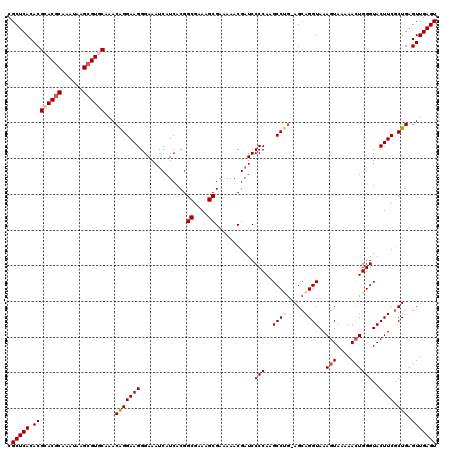
| Location | 20,734,114 – 20,734,234 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.75 |
| Mean single sequence MFE | -32.74 |
| Consensus MFE | -26.79 |
| Energy contribution | -27.99 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20734114 120 - 20766785 AGAGCCAAAUCACGACUGAUACAAAUUCGCUGCGUGAGCUAACUAUAAGUACGCUCUGUCUCGGCGAAAGAUUCUCACUCACCCUCGUACGCACACACGCCGGGACUCAAUUGGCUCUCC ((((((((.....((.(((......(((((((.(.((((..((.....))..))))...).)))))))......))).)).(((.(((........)))..)))......)))))))).. ( -35.10) >DroSec_CAF1 167643 120 - 1 AGAGCCAAAUCACGACUGAUACAAAUUCGCUGCGUGAGCUAACUAUAAGUACGCUCUGUCUCGGCGAAAGAUUCUCACUCACCCUCGUACGCACGCAGUCCGGGACUCAAUUGGCUCUCC ((((((((.....((.(((......(((((((.(.((((..((.....))..))))...).)))))))......))).))...((((.((.......)).))))......)))))))).. ( -34.60) >DroSim_CAF1 173061 120 - 1 AGAGCCAAAUCACGACUGAUACAAAUUCGCUGCGUGAGCUAACUAUAAGUACGCUCUGUCUCGGCGAAAGAUUCUCACUCACCCUCGUACGCACGCAGACCGGGACUCAAUUGGCUCUCC ((((((((.....((.(((......(((((((.(.((((..((.....))..))))...).)))))))......))).)).(((..((..(....)..)).)))......)))))))).. ( -33.80) >DroEre_CAF1 208560 120 - 1 AGAGCCAAAUCACGACUGAUACAAAUUCGCUGCGUGAGCUAACUAUAAGUACACUCUGUCUCGGCGAAAGAGUCUCACUCACCCUCGUACGCACGCACACGGGAACUCAAUGCGCUCUCC (((((((......((.(((.((...(((((((.(.(((...((.....))...)))...).)))))))...)).))).))...(((((..(....)..))))).......)).))))).. ( -27.60) >DroYak_CAF1 174314 120 - 1 AGAGCCAAAUCACGACUGAUACAAAUUCGCUGCGUGAGCUAACUAUAAGUACGCUCUGUCUCGGCGAAAGAGUCUCACUCACCCUCGAACGCACGCACACCGGGACUCAAUGCGCUCUCC (((((((.((((....)))).....(((((((.(.((((..((.....))..))))...).))))))).(((((((..((......))..(....).....)))))))..)).))))).. ( -32.60) >consensus AGAGCCAAAUCACGACUGAUACAAAUUCGCUGCGUGAGCUAACUAUAAGUACGCUCUGUCUCGGCGAAAGAUUCUCACUCACCCUCGUACGCACGCACACCGGGACUCAAUUGGCUCUCC ((((((((.....((.(((......(((((((.(.((((..((.....))..))))...).)))))))......))).)).(((.................)))......)))))))).. (-26.79 = -27.99 + 1.20)

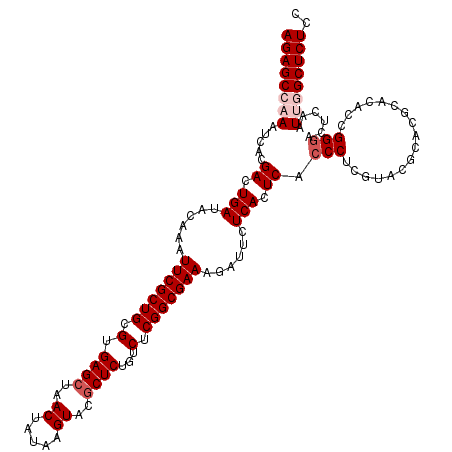
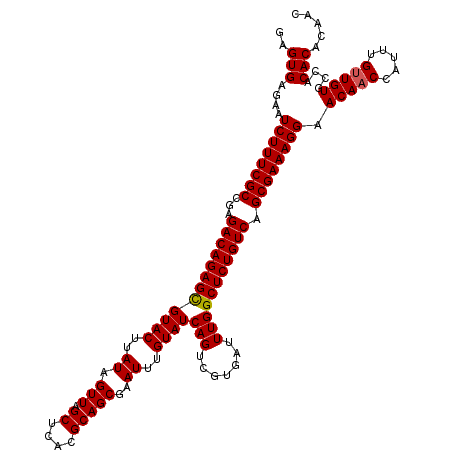
| Location | 20,734,154 – 20,734,274 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -34.96 |
| Consensus MFE | -34.14 |
| Energy contribution | -34.18 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20734154 120 + 20766785 GAGUGAGAAUCUUUCGCCGAGACAGAGCGUACUUAUAGUUAGCUCACGCAGCGAAUUUGUAUCAGUCGUGAUUUGGCUCUGUCAGCGAAAGGAACAACCAUUUGUUGUGCCACACACAAC ..(((....((((((((...((((((((((((..((.(((.((....)))))..))..))))(((.......))))))))))).)))))))).(((((.....)))))....)))..... ( -35.30) >DroSec_CAF1 167683 120 + 1 GAGUGAGAAUCUUUCGCCGAGACAGAGCGUACUUAUAGUUAGCUCACGCAGCGAAUUUGUAUCAGUCGUGAUUUGGCUCUGUCAGCGAAAGGAACAACCAUUUGUUGUGCCACACACAAC ..(((....((((((((...((((((((((((..((.(((.((....)))))..))..))))(((.......))))))))))).)))))))).(((((.....)))))....)))..... ( -35.30) >DroSim_CAF1 173101 120 + 1 GAGUGAGAAUCUUUCGCCGAGACAGAGCGUACUUAUAGUUAGCUCACGCAGCGAAUUUGUAUCAGUCGUGAUUUGGCUCUGUCAGCGAAAGGAACAACCAUUUGCUGUGCCACACACAAC ((.(((......(((((.(.....((((..((.....))..))))...).)))))......))).))(((...((((.....(((((((.((.....)).))))))).))))..)))... ( -35.80) >DroEre_CAF1 208600 120 + 1 GAGUGAGACUCUUUCGCCGAGACAGAGUGUACUUAUAGUUAGCUCACGCAGCGAAUUUGUAUCAGUCGUGAUUUGGCUCUGUCAGCGAAAGGAACAACCAUUUGUUGUGCCACACACAAC ((.(((.((...(((((.(.....((((..((.....))..))))...).)))))...)).))).))(((...((((.....(((((((.((.....)).))))))).))))..)))... ( -33.30) >DroYak_CAF1 174354 120 + 1 GAGUGAGACUCUUUCGCCGAGACAGAGCGUACUUAUAGUUAGCUCACGCAGCGAAUUUGUAUCAGUCGUGAUUUGGCUCUGUCAGCGAAAGGAACAACCAUUUGUUGUGCCACACACAAC ..(((......((((((...((((((((((((..((.(((.((....)))))..))..))))(((.......))))))))))).))))))((.(((((.....))))).)).)))..... ( -35.10) >consensus GAGUGAGAAUCUUUCGCCGAGACAGAGCGUACUUAUAGUUAGCUCACGCAGCGAAUUUGUAUCAGUCGUGAUUUGGCUCUGUCAGCGAAAGGAACAACCAUUUGUUGUGCCACACACAAC ..(((....((((((((...((((((((((((..((.(((.((....)))))..))..))))(((.......))))))))))).)))))))).(((((.....)))))....)))..... (-34.14 = -34.18 + 0.04)

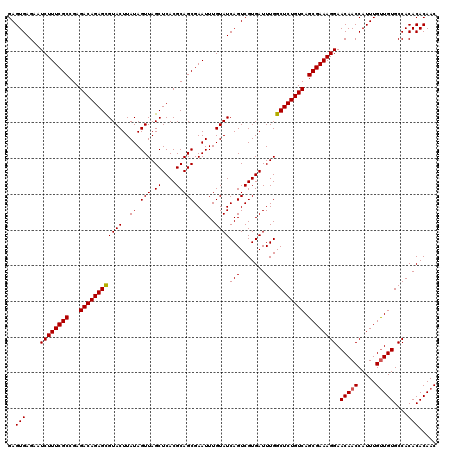
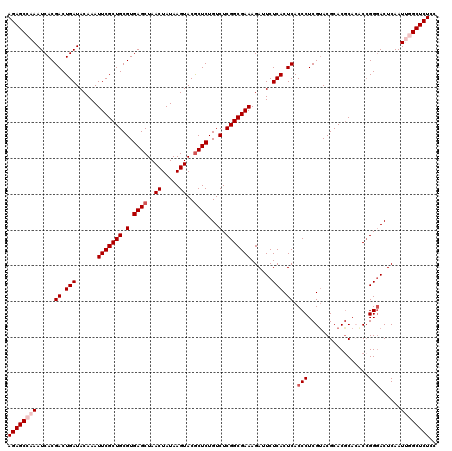
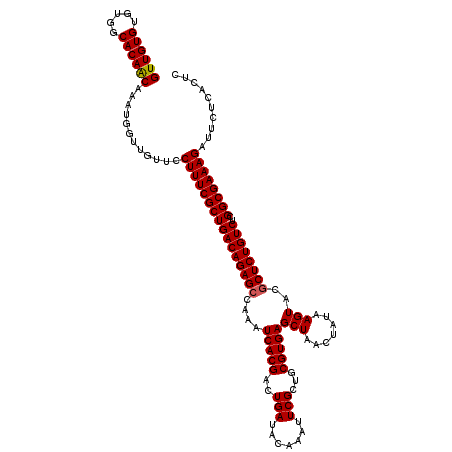
| Location | 20,734,154 – 20,734,274 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -40.04 |
| Consensus MFE | -38.76 |
| Energy contribution | -38.80 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.95 |
| SVM RNA-class probability | 0.999725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20734154 120 - 20766785 GUUGUGUGUGGCACAACAAAUGGUUGUUCCUUUCGCUGACAGAGCCAAAUCACGACUGAUACAAAUUCGCUGCGUGAGCUAACUAUAAGUACGCUCUGUCUCGGCGAAAGAUUCUCACUC ((((((.....))))))............((((((((((((((((....(((((..(((.......)))...)))))(((.......)))..))))))))..)))))))).......... ( -39.80) >DroSec_CAF1 167683 120 - 1 GUUGUGUGUGGCACAACAAAUGGUUGUUCCUUUCGCUGACAGAGCCAAAUCACGACUGAUACAAAUUCGCUGCGUGAGCUAACUAUAAGUACGCUCUGUCUCGGCGAAAGAUUCUCACUC ((((((.....))))))............((((((((((((((((....(((((..(((.......)))...)))))(((.......)))..))))))))..)))))))).......... ( -39.80) >DroSim_CAF1 173101 120 - 1 GUUGUGUGUGGCACAGCAAAUGGUUGUUCCUUUCGCUGACAGAGCCAAAUCACGACUGAUACAAAUUCGCUGCGUGAGCUAACUAUAAGUACGCUCUGUCUCGGCGAAAGAUUCUCACUC ((((((.....))))))............((((((((((((((((....(((((..(((.......)))...)))))(((.......)))..))))))))..)))))))).......... ( -39.80) >DroEre_CAF1 208600 120 - 1 GUUGUGUGUGGCACAACAAAUGGUUGUUCCUUUCGCUGACAGAGCCAAAUCACGACUGAUACAAAUUCGCUGCGUGAGCUAACUAUAAGUACACUCUGUCUCGGCGAAAGAGUCUCACUC .....(((.((((((((.....)))))..(((((((((((((((.....(((((..(((.......)))...)))))(((.......)))...)))))))..)))))))).))).))).. ( -38.60) >DroYak_CAF1 174354 120 - 1 GUUGUGUGUGGCACAACAAAUGGUUGUUCCUUUCGCUGACAGAGCCAAAUCACGACUGAUACAAAUUCGCUGCGUGAGCUAACUAUAAGUACGCUCUGUCUCGGCGAAAGAGUCUCACUC .....(((.((((((((.....)))))..((((((((((((((((....(((((..(((.......)))...)))))(((.......)))..))))))))..)))))))).))).))).. ( -42.20) >consensus GUUGUGUGUGGCACAACAAAUGGUUGUUCCUUUCGCUGACAGAGCCAAAUCACGACUGAUACAAAUUCGCUGCGUGAGCUAACUAUAAGUACGCUCUGUCUCGGCGAAAGAUUCUCACUC ((((((.....))))))............((((((((((((((((....(((((..(((.......)))...)))))(((.......)))..))))))))..)))))))).......... (-38.76 = -38.80 + 0.04)

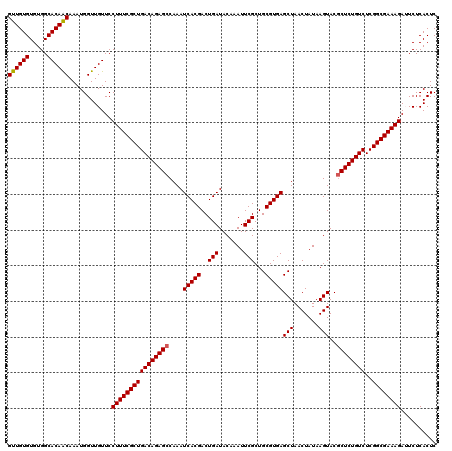
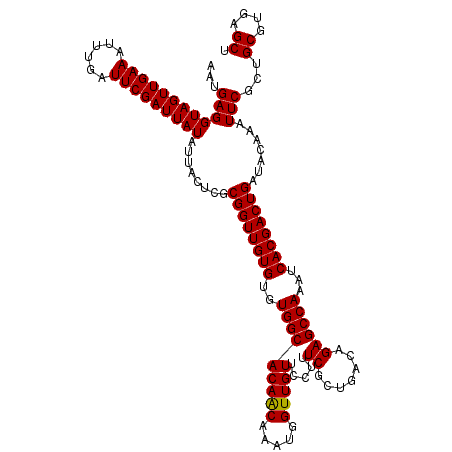
| Location | 20,734,194 – 20,734,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -33.86 |
| Consensus MFE | -32.78 |
| Energy contribution | -32.62 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20734194 120 - 20766785 AAUGAGGUAGUUGAAAUUUGAUUCGAUUAUAUUACUCGCGGUUGUGUGUGGCACAACAAAUGGUUGUUCCUUUCGCUGACAGAGCCAAAUCACGACUGAUACAAAUUCGCUGCGUGAGCU ...((((((((((((......)))))))))....))).((((((((..(((((((((.....))))).....((.......))))))...)))))))).............((....)). ( -33.10) >DroSec_CAF1 167723 120 - 1 AAUGAGGUAGUUGAAGUUUGAUUCGAUUAUAUUACACGCGGUUGUGUGUGGCACAACAAAUGGUUGUUCCUUUCGCUGACAGAGCCAAAUCACGACUGAUACAAAUUCGCUGCGUGAGCU ..(((.(((((((((......))))))))).)))((((((((((((..(((((((((.....))))).....((.......))))))...)))))))((.......))...))))).... ( -34.40) >DroSim_CAF1 173141 120 - 1 AAUGAGGUAGUUGAAGUUUGAUUCGAUUAUAUUACACGCGGUUGUGUGUGGCACAGCAAAUGGUUGUUCCUUUCGCUGACAGAGCCAAAUCACGACUGAUACAAAUUCGCUGCGUGAGCU ..(((.(((((((((......))))))))).)))((((((((((((..((((.((((.((.((.....)).)).)))).....))))...)))))))((.......))...))))).... ( -36.30) >DroEre_CAF1 208640 120 - 1 AAUGAGGUAGUUGAAAUUUGAUUCGAUUAUAUUAUUCGCGGUUGUGUGUGGCACAACAAAUGGUUGUUCCUUUCGCUGACAGAGCCAAAUCACGACUGAUACAAAUUCGCUGCGUGAGCU (((((.(((((((((......))))))))).)))))..((((((((..(((((((((.....))))).....((.......))))))...)))))))).............((....)). ( -33.20) >DroYak_CAF1 174394 120 - 1 AAUGAGGUAGUUGAAAUUUGAUUCGAUUAUAUUUUUCGCGGUUGUGUGUGGCACAACAAAUGGUUGUUCCUUUCGCUGACAGAGCCAAAUCACGACUGAUACAAAUUCGCUGCGUGAGCU ...((((((((((((......)))))))))........((((((((..(((((((((.....))))).....((.......))))))...)))))))).......)))...((....)). ( -32.30) >consensus AAUGAGGUAGUUGAAAUUUGAUUCGAUUAUAUUACUCGCGGUUGUGUGUGGCACAACAAAUGGUUGUUCCUUUCGCUGACAGAGCCAAAUCACGACUGAUACAAAUUCGCUGCGUGAGCU ...((((((((((((......)))))))))........((((((((..(((((((((.....))))).....((.......))))))...)))))))).......)))...((....)). (-32.78 = -32.62 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:01 2006