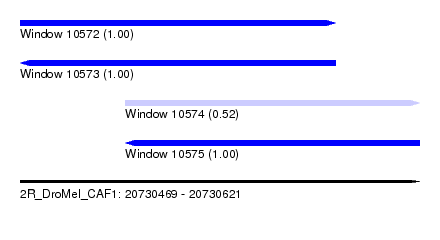
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,730,469 – 20,730,621 |
| Length | 152 |
| Max. P | 0.998480 |

| Location | 20,730,469 – 20,730,589 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -33.78 |
| Consensus MFE | -32.06 |
| Energy contribution | -32.06 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.995911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20730469 120 + 20766785 UUUUUCAGACAAAUAAUCCAGCCUUAAGCAUGGUGAUUAAGCUUGAUCCCCUACCAAGGGGCGUAAUAUUGACGGAUUUUCCUUAAAUCCCUCUGUUAAUCUCCGGCUUAGAGCGCGACG ....................((.((((((.(((.((((((.......(((((....))))).........((.((((((.....)))))).))..)))))).))))))))).))...... ( -32.00) >DroSec_CAF1 163967 120 + 1 UUUUUCAGACAAAUAAUCCAGCCUUAAGCAUGGCGAUUAAGCUUGAUCCCCUACCAAGGGGCGUAAUAUUGACGGAUUUUCCUUAAAUCCCUCUGUUAAUCUCCGGCUUAGAGCGCGACG ....................((.((((((.(((.((((((.......(((((....))))).........((.((((((.....)))))).))..)))))).))))))))).))...... ( -31.70) >DroSim_CAF1 169389 120 + 1 UUUUUCAGACAAAUAAUCCAGCCUUAAGCAUGGCGAUUAAGCUUGAUCCCCUACCAAGGGGCGUAAUAUUGACGGAUUUUCCUUAAAUCCCUCUGUUAAUCUCCGGCUUAGAGCGCGACG ....................((.((((((.(((.((((((.......(((((....))))).........((.((((((.....)))))).))..)))))).))))))))).))...... ( -31.70) >DroEre_CAF1 204888 120 + 1 UUUUUCAGACAAAUAAUCCAGCCUUAAGCAUGGUGAUUAAGCUUGAUCCCCUACCAAGGGGCGUAAUAUUGACGGAUUUUCCUUAAAUCCCUCGCUUAAUCUCCGGCUUAGAGCGCGACG ....................((.((((((.(((.((((((((.....(((((....))))).........((.((((((.....)))))).)))))))))).))))))))).))...... ( -39.00) >DroYak_CAF1 170549 120 + 1 UUUUUCAGACAAAUAAUCCAGCCUUAAGCAUGGUGAUUAAGCUUGAUCCCCUACCAAGGGGCGUAAUAUUGACGGAUUUUCCUUAAAUCCCUCGGUUAAUCUCCGGCUUAGAGCGCGACG ....................((.((((((.(((.((((((.(.....(((((....))))).........((.((((((.....)))))).))).)))))).))))))))).))...... ( -34.50) >consensus UUUUUCAGACAAAUAAUCCAGCCUUAAGCAUGGUGAUUAAGCUUGAUCCCCUACCAAGGGGCGUAAUAUUGACGGAUUUUCCUUAAAUCCCUCUGUUAAUCUCCGGCUUAGAGCGCGACG ....................((.((((((.(((.((((((.......(((((....))))).........((.((((((.....)))))).))..)))))).))))))))).))...... (-32.06 = -32.06 + -0.00)


| Location | 20,730,469 – 20,730,589 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -36.96 |
| Consensus MFE | -35.30 |
| Energy contribution | -35.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20730469 120 - 20766785 CGUCGCGCUCUAAGCCGGAGAUUAACAGAGGGAUUUAAGGAAAAUCCGUCAAUAUUACGCCCCUUGGUAGGGGAUCAAGCUUAAUCACCAUGCUUAAGGCUGGAUUAUUUGUCUGAAAAA ......((..(((((.((.(((((((.((.((((((.....)))))).)).........(((((....))))).....).)))))).))..)))))..)).((((.....))))...... ( -35.20) >DroSec_CAF1 163967 120 - 1 CGUCGCGCUCUAAGCCGGAGAUUAACAGAGGGAUUUAAGGAAAAUCCGUCAAUAUUACGCCCCUUGGUAGGGGAUCAAGCUUAAUCGCCAUGCUUAAGGCUGGAUUAUUUGUCUGAAAAA ......((..(((((.((.(((((((.((.((((((.....)))))).)).........(((((....))))).....).)))))).))..)))))..)).((((.....))))...... ( -35.70) >DroSim_CAF1 169389 120 - 1 CGUCGCGCUCUAAGCCGGAGAUUAACAGAGGGAUUUAAGGAAAAUCCGUCAAUAUUACGCCCCUUGGUAGGGGAUCAAGCUUAAUCGCCAUGCUUAAGGCUGGAUUAUUUGUCUGAAAAA ......((..(((((.((.(((((((.((.((((((.....)))))).)).........(((((....))))).....).)))))).))..)))))..)).((((.....))))...... ( -35.70) >DroEre_CAF1 204888 120 - 1 CGUCGCGCUCUAAGCCGGAGAUUAAGCGAGGGAUUUAAGGAAAAUCCGUCAAUAUUACGCCCCUUGGUAGGGGAUCAAGCUUAAUCACCAUGCUUAAGGCUGGAUUAUUUGUCUGAAAAA ......((..(((((.((.((((((((((.((((((.....)))))).)).........(((((....))))).....)))))))).))..)))))..)).((((.....))))...... ( -42.40) >DroYak_CAF1 170549 120 - 1 CGUCGCGCUCUAAGCCGGAGAUUAACCGAGGGAUUUAAGGAAAAUCCGUCAAUAUUACGCCCCUUGGUAGGGGAUCAAGCUUAAUCACCAUGCUUAAGGCUGGAUUAUUUGUCUGAAAAA ......((..(((((.((.((((((.(((.((((((.....)))))).)).........(((((....))))).....).)))))).))..)))))..)).((((.....))))...... ( -35.80) >consensus CGUCGCGCUCUAAGCCGGAGAUUAACAGAGGGAUUUAAGGAAAAUCCGUCAAUAUUACGCCCCUUGGUAGGGGAUCAAGCUUAAUCACCAUGCUUAAGGCUGGAUUAUUUGUCUGAAAAA ......((..(((((.((.((((((..((.((((((.....)))))).)).........(((((....))))).......)))))).))..)))))..)).((((.....))))...... (-35.30 = -35.30 + 0.00)

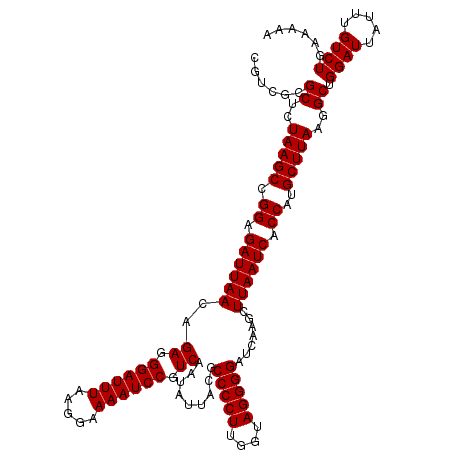
| Location | 20,730,509 – 20,730,621 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.29 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -26.68 |
| Energy contribution | -26.84 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.91 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20730509 112 + 20766785 GCUUGAUCCCCUACCAAGGGGCGUAAUAUUGACGGAUUUUCCUUAAAUCCCUCUGUUAAUCUCCGGCUUAGAGCGCGACGCGUUU-UUUCGCGACUCCGCCUGCAUUGUUUUU------- ((..((((((((....)))))..(((((..((.((((((.....)))))).))))))))))...(((...((((((((.......-..))))).))).))).)).........------- ( -30.00) >DroSec_CAF1 164007 112 + 1 GCUUGAUCCCCUACCAAGGGGCGUAAUAUUGACGGAUUUUCCUUAAAUCCCUCUGUUAAUCUCCGGCUUAGAGCGCGACGCGUUU-UUUCGCGACUCCGCCUGCAUUGUUUUU------- ((..((((((((....)))))..(((((..((.((((((.....)))))).))))))))))...(((...((((((((.......-..))))).))).))).)).........------- ( -30.00) >DroSim_CAF1 169429 112 + 1 GCUUGAUCCCCUACCAAGGGGCGUAAUAUUGACGGAUUUUCCUUAAAUCCCUCUGUUAAUCUCCGGCUUAGAGCGCGACGCGUUU-UUUCGCGACUCCGCCUGCAUUGUUUUU------- ((..((((((((....)))))..(((((..((.((((((.....)))))).))))))))))...(((...((((((((.......-..))))).))).))).)).........------- ( -30.00) >DroEre_CAF1 204928 120 + 1 GCUUGAUCCCCUACCAAGGGGCGUAAUAUUGACGGAUUUUCCUUAAAUCCCUCGCUUAAUCUCCGGCUUAGAGCGCGACGCGUUUUUUUCGCGACACCGUCUGCAUUGUUUUUUUUUUUU ((..(((.((((....))))((((......((.((((((.....)))))).))(((........))).....))))..((((.......)))).....))).))................ ( -27.40) >DroYak_CAF1 170589 111 + 1 GCUUGAUCCCCUACCAAGGGGCGUAAUAUUGACGGAUUUUCCUUAAAUCCCUCGGUUAAUCUCCGGCUUAGAGCGCGACGCGUUUUUUUCGCGACUCCUUCUGCAUUGUUU--------- (((.((.(((((....))))).(....(((((.((((((.....)))))).)))))....))).)))...((((((((..........))))).)))..............--------- ( -28.70) >consensus GCUUGAUCCCCUACCAAGGGGCGUAAUAUUGACGGAUUUUCCUUAAAUCCCUCUGUUAAUCUCCGGCUUAGAGCGCGACGCGUUU_UUUCGCGACUCCGCCUGCAUUGUUUUU_______ ((..((((((((....))))).........((.((((((.....)))))).)).....)))...(((...((((((((..........))))).))).))).))................ (-26.68 = -26.84 + 0.16)

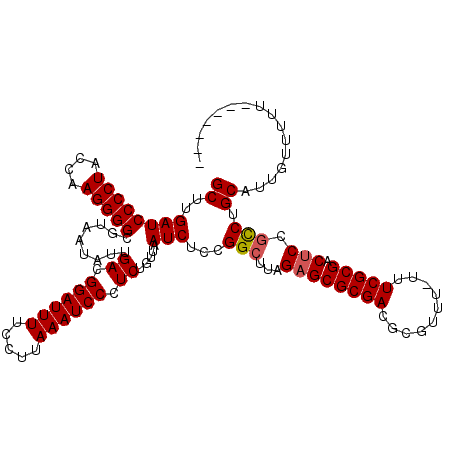
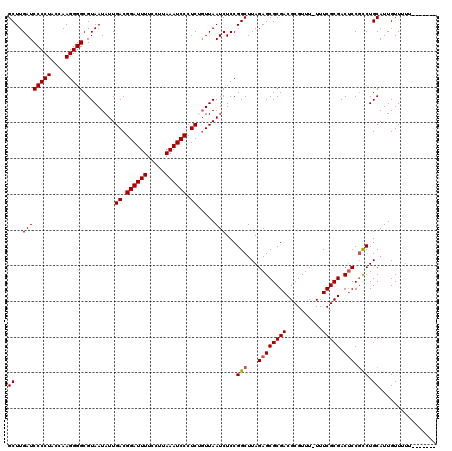
| Location | 20,730,509 – 20,730,621 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.29 |
| Mean single sequence MFE | -33.36 |
| Consensus MFE | -30.26 |
| Energy contribution | -31.06 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.998480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20730509 112 - 20766785 -------AAAAACAAUGCAGGCGGAGUCGCGAAA-AAACGCGUCGCGCUCUAAGCCGGAGAUUAACAGAGGGAUUUAAGGAAAAUCCGUCAAUAUUACGCCCCUUGGUAGGGGAUCAAGC -------.........((.(((((((.(((((..-.......)))))))))..)))...(((.....((.((((((.....)))))).)).........(((((....))))))))..)) ( -34.80) >DroSec_CAF1 164007 112 - 1 -------AAAAACAAUGCAGGCGGAGUCGCGAAA-AAACGCGUCGCGCUCUAAGCCGGAGAUUAACAGAGGGAUUUAAGGAAAAUCCGUCAAUAUUACGCCCCUUGGUAGGGGAUCAAGC -------.........((.(((((((.(((((..-.......)))))))))..)))...(((.....((.((((((.....)))))).)).........(((((....))))))))..)) ( -34.80) >DroSim_CAF1 169429 112 - 1 -------AAAAACAAUGCAGGCGGAGUCGCGAAA-AAACGCGUCGCGCUCUAAGCCGGAGAUUAACAGAGGGAUUUAAGGAAAAUCCGUCAAUAUUACGCCCCUUGGUAGGGGAUCAAGC -------.........((.(((((((.(((((..-.......)))))))))..)))...(((.....((.((((((.....)))))).)).........(((((....))))))))..)) ( -34.80) >DroEre_CAF1 204928 120 - 1 AAAAAAAAAAAACAAUGCAGACGGUGUCGCGAAAAAAACGCGUCGCGCUCUAAGCCGGAGAUUAAGCGAGGGAUUUAAGGAAAAUCCGUCAAUAUUACGCCCCUUGGUAGGGGAUCAAGC ................((.....(((.((((.......)))).))).((((.....)))).....))((.((((((.....)))))).)).........(((((....)))))....... ( -32.10) >DroYak_CAF1 170589 111 - 1 ---------AAACAAUGCAGAAGGAGUCGCGAAAAAAACGCGUCGCGCUCUAAGCCGGAGAUUAACCGAGGGAUUUAAGGAAAAUCCGUCAAUAUUACGCCCCUUGGUAGGGGAUCAAGC ---------.............((((.(((((..........)))))))))..((..((........((.((((((.....)))))).)).........(((((....))))).))..)) ( -30.30) >consensus _______AAAAACAAUGCAGGCGGAGUCGCGAAA_AAACGCGUCGCGCUCUAAGCCGGAGAUUAACAGAGGGAUUUAAGGAAAAUCCGUCAAUAUUACGCCCCUUGGUAGGGGAUCAAGC ................((.(((((((.(((((..........)))))))))..)))...(((.....((.((((((.....)))))).)).........(((((....))))))))..)) (-30.26 = -31.06 + 0.80)


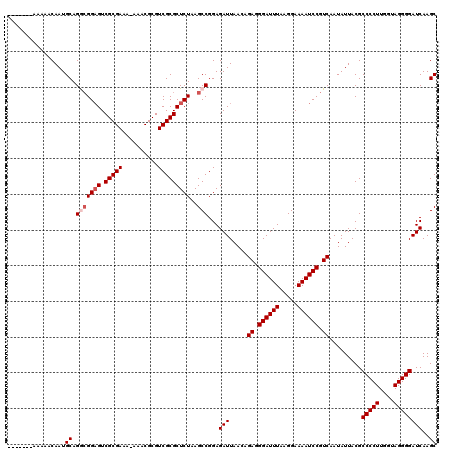
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:56 2006