
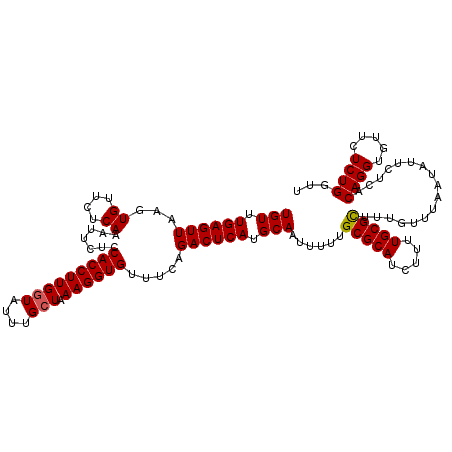
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,723,475 – 20,723,595 |
| Length | 120 |
| Max. P | 0.947510 |

| Location | 20,723,475 – 20,723,595 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -24.68 |
| Energy contribution | -24.72 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20723475 120 + 20766785 UGUUUGAGUUAAGUGUUCUCAAAUUCUCCACCUUGGUAUUUGCUAAAGGUGUUUCGGACUCAUGCAAUUUUUGCGCAUCUUUUGCGCUUUGUUUAAUAUUCUCACAGGUGUUCUCUGGUU (((.((((((...((....)).......(((((((((....))).)))))).....)))))).)))......(((((.....))))).................((((.....))))... ( -26.20) >DroSec_CAF1 156865 120 + 1 UGUUUGAGUUAAGUGUUCUCAAAUUCUCCACCUUGGUAUUUGCUAAAGGUGUUUCAGACUCAUGCAAUUUUUGCGCAUCUUUUGCGCUUUGUUUAAUAUUCUCACAGGUGUUCUCUGGUU (((.((((((...((....)).......(((((((((....))).)))))).....)))))).)))......(((((.....))))).................((((.....))))... ( -26.10) >DroSim_CAF1 161953 120 + 1 UGUUUGAGUUAAGUGUUCUCAAAUUCUCCACCUUGAUAUUUGCUAAAGGUGUUUCAGACUCAUGCAAUUUUUGCGCAUCUUUUGCGCUUUGUUUAAUAUUCUCACAGGUGUUGUCUGGUU .(((((((.........)))))))....((((((...........))))))..((((((....((((.....(((((.....))))).))))..((((((......)))))))))))).. ( -27.40) >DroEre_CAF1 197940 120 + 1 UGUUUGAGUUAAGUGUUCUCAAAUUCUCCACCUUGGUAUUUGCUAAAGGUGUUUCAGACUCAUGCAAUUUUUGCGCAUCUUUUGCGCUUUGUUUAAUAUUCUCACAGGUGUUCUCUGGUU (((.((((((...((....)).......(((((((((....))).)))))).....)))))).)))......(((((.....))))).................((((.....))))... ( -26.10) >DroYak_CAF1 163172 120 + 1 UGUUUGAGUUAAGUGUUCUCAUAUUCUCCACCUUGGUAUUUGCUAAAGGUGUUUCAGACUCAUGCAAUUUUUGCGCAUCUUUUGCGUUUUGUUUAAUAUUCUCACAGGUGUUCUCUGGCU (((.((((((..(((....)))......(((((((((....))).)))))).....)))))).)))......(((((.....))))).................((((.....))))... ( -25.20) >consensus UGUUUGAGUUAAGUGUUCUCAAAUUCUCCACCUUGGUAUUUGCUAAAGGUGUUUCAGACUCAUGCAAUUUUUGCGCAUCUUUUGCGCUUUGUUUAAUAUUCUCACAGGUGUUCUCUGGUU (((.((((((...((....)).......(((((((((....))).)))))).....)))))).)))......(((((.....))))).................((((.....))))... (-24.68 = -24.72 + 0.04)

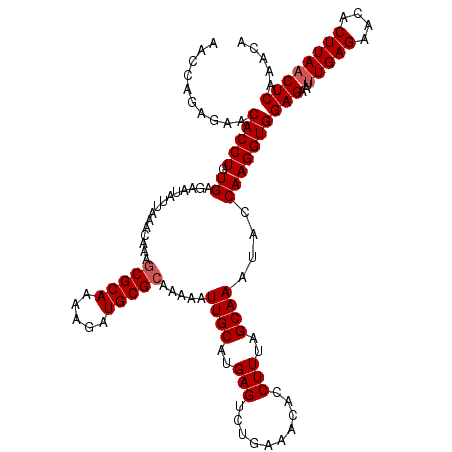
| Location | 20,723,475 – 20,723,595 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -21.96 |
| Consensus MFE | -19.62 |
| Energy contribution | -20.02 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20723475 120 - 20766785 AACCAGAGAACACCUGUGAGAAUAUUAAACAAAGCGCAAAAGAUGCGCAAAAAUUGCAUGAGUCCGAAACACCUUUAGCAAAUACCAAGGUGGAGAAUUUGAGAACACUUAACUCAAACA .....(((..(((((.((...............(((((.....))))).....((((..(((..........)))..))))....)))))))......(((((....))))))))..... ( -22.50) >DroSec_CAF1 156865 120 - 1 AACCAGAGAACACCUGUGAGAAUAUUAAACAAAGCGCAAAAGAUGCGCAAAAAUUGCAUGAGUCUGAAACACCUUUAGCAAAUACCAAGGUGGAGAAUUUGAGAACACUUAACUCAAACA .....(((..(((((.((...............(((((.....))))).....((((..(((..........)))..))))....)))))))......(((((....))))))))..... ( -22.50) >DroSim_CAF1 161953 120 - 1 AACCAGACAACACCUGUGAGAAUAUUAAACAAAGCGCAAAAGAUGCGCAAAAAUUGCAUGAGUCUGAAACACCUUUAGCAAAUAUCAAGGUGGAGAAUUUGAGAACACUUAACUCAAACA ...(((((..((....))...............(((((.....))))).............)))))...((((((...........)))))).....((((((.........)))))).. ( -24.30) >DroEre_CAF1 197940 120 - 1 AACCAGAGAACACCUGUGAGAAUAUUAAACAAAGCGCAAAAGAUGCGCAAAAAUUGCAUGAGUCUGAAACACCUUUAGCAAAUACCAAGGUGGAGAAUUUGAGAACACUUAACUCAAACA .....(((..(((((.((...............(((((.....))))).....((((..(((..........)))..))))....)))))))......(((((....))))))))..... ( -22.50) >DroYak_CAF1 163172 120 - 1 AGCCAGAGAACACCUGUGAGAAUAUUAAACAAAACGCAAAAGAUGCGCAAAAAUUGCAUGAGUCUGAAACACCUUUAGCAAAUACCAAGGUGGAGAAUAUGAGAACACUUAACUCAAACA .((((.((.....)).))................((((.....))))........)).(((((......((((((...........)))))).......((((....))))))))).... ( -18.00) >consensus AACCAGAGAACACCUGUGAGAAUAUUAAACAAAGCGCAAAAGAUGCGCAAAAAUUGCAUGAGUCUGAAACACCUUUAGCAAAUACCAAGGUGGAGAAUUUGAGAACACUUAACUCAAACA ..........(((((.((...............(((((.....))))).....((((..(((..........)))..))))....)))))))(((...(((((....))))))))..... (-19.62 = -20.02 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:45 2006