
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,723,341 – 20,723,450 |
| Length | 109 |
| Max. P | 0.959156 |

| Location | 20,723,341 – 20,723,450 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.25 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -22.80 |
| Energy contribution | -22.36 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
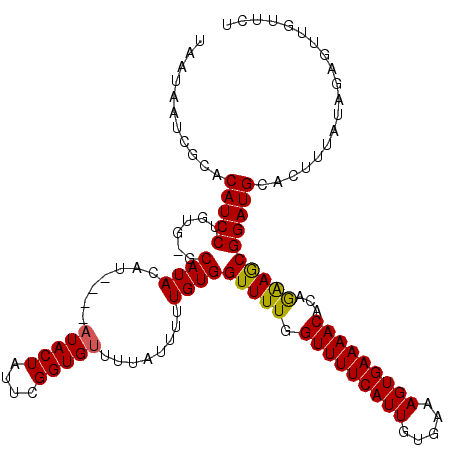
>2R_DroMel_CAF1 20723341 109 + 20766785 UAAUAAUCGCACAUCCUGUGCGCAUACA------UACUAUUCGGUGUU-----UUUGUGGUUUUGGUUUUCAUUGUGAAAGUGAAAACACAGAAGCGGAUGCACUUUAUAGAGUUGUUUU .(((((((((((.....)))))......------..((((..(((((.-----(((((..((((((((((((((.....))))))))).)))))))))).)))))..)))).)))))).. ( -32.40) >DroSec_CAF1 156724 116 + 1 UGAUAAUCGCACAUCCUGUGAGCAUACAU----AUACUAUUCGGUGUUUUAUUUUUGUGGUUUUGGUUUUCAUUGUGAAAGUGAAAACACAGAAGCGGAUGCACUUUAUAGAGUUCUUCU ......(((((.....)))))........----...((((..(((((((..((((((((.((((..(((((.....)))))..))))))))))))..)).)))))..))))......... ( -26.30) >DroSim_CAF1 161808 120 + 1 UAAUAAUCGCACAUCCUGUGAGCAUACAUAUGUAUACUAUUCGGUGUUUUAUUUUUGUGGUUUUGGUUUUCAUUGUGAAAGUGAAAACACAGAAGCGGAUGCACUUUAUAGAGUUCUUCU ...................((((((((....)))).((((..(((((((..((((((((.((((..(((((.....)))))..))))))))))))..)).)))))..)))).)))).... ( -28.00) >DroEre_CAF1 197786 114 + 1 UUAUAAUCGCACAUCCUGUG--CAUACAU----AUACUAUUUGGUGUUUUGUUUUUGUGGUUUUGGUUUUCAUUGUAAAAGUGAAAACACAAAAACGGAUGCACUUUAUACAUAUGAUCA ........((((.....)))--)...(((----((..(((..((((((((((((((((.......(((((((((.....)))))))))))))))))))).)))))..))).))))).... ( -31.81) >DroYak_CAF1 163018 114 + 1 UAAUAAUCGCACAUCCUGUG--CAUACAU----AUACUAUUUGGUGUUCUGUUUUUGUGGUUUUGGUUUUCAUUGUGAAAGUGAAAACACAAGAACGGAUGCACUUUAUACAGCUGAUCG .....(((((((.....)))--)......----....(((..(((((((((((((((((.((((..(((((.....)))))..)))))))))))))))).)))))..))).....))).. ( -33.10) >consensus UAAUAAUCGCACAUCCUGUG_GCAUACAU____AUACUAUUCGGUGUUUUAUUUUUGUGGUUUUGGUUUUCAUUGUGAAAGUGAAAACACAGAAGCGGAUGCACUUUAUAGAGUUGUUCU ...........(((((......((((.......(((((....)))))........))))(((((.(((((((((.....)))))))))...))))))))))................... (-22.80 = -22.36 + -0.44)

| Location | 20,723,341 – 20,723,450 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.25 |
| Mean single sequence MFE | -22.39 |
| Consensus MFE | -17.88 |
| Energy contribution | -17.88 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
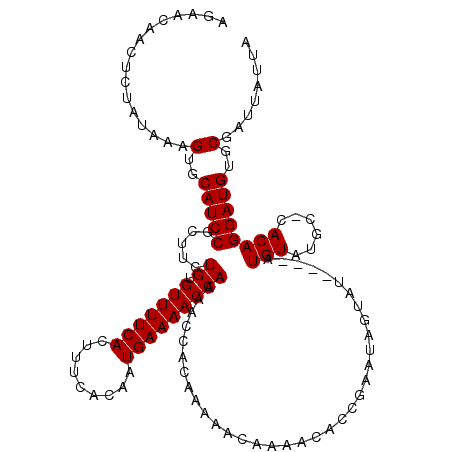
>2R_DroMel_CAF1 20723341 109 - 20766785 AAAACAACUCUAUAAAGUGCAUCCGCUUCUGUGUUUUCACUUUCACAAUGAAAACCAAAACCACAAA-----AACACCGAAUAGUA------UGUAUGCGCACAGGAUGUGCGAUUAUUA ................(..(((((..(((.(((((((...((((.....))))............))-----))))).)))..((.------((....)).)).)))))..)........ ( -18.76) >DroSec_CAF1 156724 116 - 1 AGAAGAACUCUAUAAAGUGCAUCCGCUUCUGUGUUUUCACUUUCACAAUGAAAACCAAAACCACAAAAAUAAAACACCGAAUAGUAU----AUGUAUGCUCACAGGAUGUGCGAUUAUCA ................(..(((((.....((.(((((((.........))))))))).........................(((((----....)))))....)))))..)........ ( -19.50) >DroSim_CAF1 161808 120 - 1 AGAAGAACUCUAUAAAGUGCAUCCGCUUCUGUGUUUUCACUUUCACAAUGAAAACCAAAACCACAAAAAUAAAACACCGAAUAGUAUACAUAUGUAUGCUCACAGGAUGUGCGAUUAUUA ................(..(((((.....((.(((((((.........))))))))).........................(((((((....)))))))....)))))..)........ ( -24.30) >DroEre_CAF1 197786 114 - 1 UGAUCAUAUGUAUAAAGUGCAUCCGUUUUUGUGUUUUCACUUUUACAAUGAAAACCAAAACCACAAAAACAAAACACCAAAUAGUAU----AUGUAUG--CACAGGAUGUGCGAUUAUAA ..........(((((.(..((((((((((((((((((((.........))))))).......)))))))).............((((----....)))--)...)))))..)..))))). ( -25.91) >DroYak_CAF1 163018 114 - 1 CGAUCAGCUGUAUAAAGUGCAUCCGUUCUUGUGUUUUCACUUUCACAAUGAAAACCAAAACCACAAAAACAGAACACCAAAUAGUAU----AUGUAUG--CACAGGAUGUGCGAUUAUUA ................(..((((((((((((((((((...((((.....))))...)))).)))).....)))))........((((----....)))--)...)))))..)........ ( -23.50) >consensus AGAACAACUCUAUAAAGUGCAUCCGCUUCUGUGUUUUCACUUUCACAAUGAAAACCAAAACCACAAAAACAAAACACCGAAUAGUAU____AUGUAUGC_CACAGGAUGUGCGAUUAUUA ................(..(((((.....((.(((((((.........)))))))))...................................(((......))))))))..)........ (-17.88 = -17.88 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:43 2006