| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,722,662 – 20,722,812 |
| Length | 150 |
| Max. P | 0.853131 |

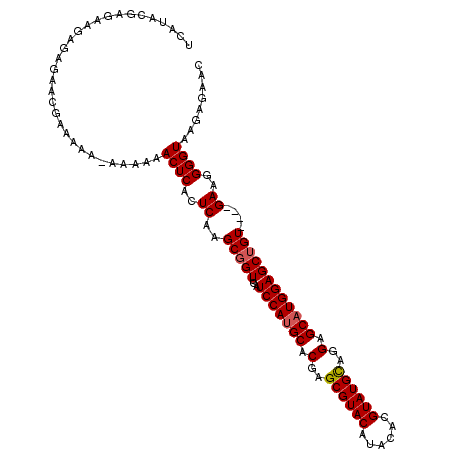
| Location | 20,722,662 – 20,722,772 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 92.88 |
| Mean single sequence MFE | -30.19 |
| Consensus MFE | -25.02 |
| Energy contribution | -25.47 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20722662 110 + 20766785 UCAUACGAGAAGAGAGAACGAAAA----AAAACUCACUCAAGCGGUUCAUCCACGCACGAGCGUACAUACACGUAUGUAGGAGCAUGGAGCUGUGUAUGAAGGGGUAAGAGAAC (((((((((..(((..........----....))).)))..(((((((.(((((((....))))(((((....))))).))).....))))))))))))).............. ( -29.34) >DroSec_CAF1 156006 109 + 1 UCAUACGAGAAGAGAGAACGAAAAA-AAAAAACUCACUCAAGCCGUUCAUCCAUGCACGAGCGUACAUACACGUAUGCAGGAGCAUGGAGCUGU----GAAGGGGUAAGAGAAC ((.((((((..(((...........-......))).)))...((.(((((((((((.(..((((((......))))))..).)))))))....)----))).))))).)).... ( -30.33) >DroSim_CAF1 161088 110 + 1 UCAUACGAGAAGAGAGAACGAAAAAAAAAAAACUCACUCAAGCGGUUCAUCCAUGCACAAGCGUACAUACACGUAUGCAGGAGCAUGGAGCUGU----GAAGGGGUAAGAGAAC ................................(((((((..(((((...(((((((.(..((((((......))))))..).))))))))))))----....))))..)))... ( -30.90) >consensus UCAUACGAGAAGAGAGAACGAAAAA_AAAAAACUCACUCAAGCGGUUCAUCCAUGCACGAGCGUACAUACACGUAUGCAGGAGCAUGGAGCUGU____GAAGGGGUAAGAGAAC ...............................((((..((..(((((...(((((((.(..((((((......))))))..).))))))))))))....))..))))........ (-25.02 = -25.47 + 0.45)


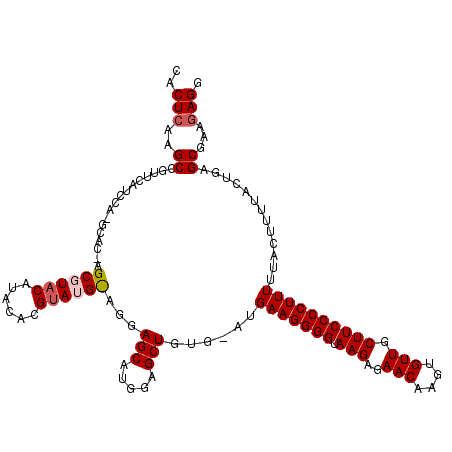
| Location | 20,722,692 – 20,722,812 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.94 |
| Mean single sequence MFE | -35.64 |
| Consensus MFE | -20.82 |
| Energy contribution | -21.74 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20722692 120 + 20766785 CACUCAAGCGGUUCAUCCACGCACGAGCGUACAUACACGUAUGUAGGAGCAUGGAGCUGUGUAUGAAGGGGUAAGAGAACAAGUGUUUCUUCCCCUUUUUACUUUCACUGAGCGAAGAGG (.((((.(((((((.(((((((....))))(((((....))))).))).....)))))))(((.(((((((.((((((.(....)))))))))))))).)))......)))).)...... ( -41.50) >DroSec_CAF1 156039 116 + 1 CACUCAAGCCGUUCAUCCAUGCACGAGCGUACAUACACGUAUGCAGGAGCAUGGAGCUGU----GAAGGGGUAAGAGAACAAGUGUUGCUUCCCCUUUUUACUUUUACUGAGCGAAGAGG ..(((....(((((((((((((.(..((((((......))))))..).))))))).....----(((((((.(((..(((....))).))))))))))..........))))))..))). ( -41.60) >DroSim_CAF1 161122 116 + 1 CACUCAAGCGGUUCAUCCAUGCACAAGCGUACAUACACGUAUGCAGGAGCAUGGAGCUGU----GAAGGGGUAAGAGAACAAGUGUUGCUUCCCCUUUUUACUUUUACUGAGCGAAGAGG (.((((.(((((...(((((((.(..((((((......))))))..).))))))))))))----(((((((.(((..(((....))).))))))))))..........)))).)...... ( -41.50) >DroEre_CAF1 197163 98 + 1 CACUCAAGCC------------------GUACACACACGUA----CGAGCAUGGAGCUGAGAAUGAAGGGGUAAGAGAACAAGUGUUGCUUCCCCUUUUUACUUUUACGGAGCGAAAAGG (((((..(((------------------((((......)))----)).))...))).)).....(((((((.(((..(((....))).))))))))))...(((((.((...))))))). ( -27.20) >DroYak_CAF1 162356 98 + 1 CACUCAAGCC------------------GUACACACACGGA----CGAGCAUGGAGCUGAGAAUGAAGGGGUAAGAGAACAAGUGUUGCUUCCCCUUUUUACUUUUACUGAGCGAAGAGU .((((..(((------------------((.(......).)----)).)).....(((.((((.(((((((.(((..(((....))).))))))))))......)).)).)))...)))) ( -26.40) >consensus CACUCAAGCCGUUCAUCCA_GCAC_AGCGUACAUACACGUAUG_AGGAGCAUGGAGCUGUG_AUGAAGGGGUAAGAGAACAAGUGUUGCUUCCCCUUUUUACUUUUACUGAGCGAAGAGG ..(((..((.................((((((......))))))...(((.....)))......(((((((.(((..(((....))).)))))))))).............))...))). (-20.82 = -21.74 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:40 2006