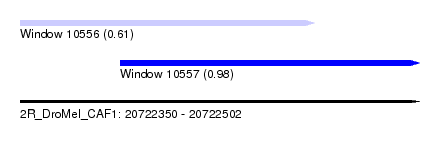
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,722,350 – 20,722,502 |
| Length | 152 |
| Max. P | 0.980260 |

| Location | 20,722,350 – 20,722,462 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -17.82 |
| Consensus MFE | -11.56 |
| Energy contribution | -11.04 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20722350 112 + 20766785 U--AAGCAAUACGCCAUUUCAUAGGUAAUAUUGAUCAAACUCCGCCUAUU--UUUUACUCAUAACAGUAUACACCCAAAUACAU----ACAUACACACAUGUGUGUAUUUCAGAAAUAUA .--..((.....)).(((((((((((.................)))))).--...((((......)))).......((((((((----((((......))))))))))))..)))))... ( -18.13) >DroSec_CAF1 155696 112 + 1 U--AGGCAGUACGCCAUUUCAUAGGUAAUAUUGAUCAAACUCCGCCUUUU--UCUUACUCAUAUCAGUAUACACACAAAUACAU----ACAUACACACAUGUGUGUAUUUCAGAAAUAUA .--.(((.....)))(((((....(((.(((((((...............--..........))))))))))....((((((((----((((......))))))))))))..)))))... ( -22.61) >DroSim_CAF1 160774 116 + 1 U--AGGCAAUACGCCAUUUCAUAGGUAAUAUUGAUCAAACUCCGCCUUUU--UCUUACUCAUAUCAGUAUACACACAAAUACAUAUGUACAUACACACAUGUGUGUAUUUCAGAAAUAUA .--.(((.....)))(((((....(((.(((((((...............--..........))))))))))......((((((((((........))))))))))......)))))... ( -21.21) >DroEre_CAF1 196825 109 + 1 UAAUAGCAGUACGCCAUUUCAUAGGUAAUAUCGAUCAAACUCCGCCUUUUUAUUUGACUCAUA---CAGUGCAUACAAAUACAU----UC----AUACAUGUGUGUAUUUCAGAAAUAUA ........(((((((........)))......(((((((.............))))).))...---..))))....((((((((----.(----(....)).)))))))).......... ( -13.92) >DroYak_CAF1 162011 112 + 1 UCAAAGCAGUACACCAUUUCAUAGGUAAUAUCGAUCAAACUCCGCCUUCCUUUUUUACUCAUAUCACAGUACGCACAAAUACAA----AC----AUACAUGUGUGUAUUUUAGAAAUAUA ...............(((((..((((.................))))....................((((((((((..((...----..----.))..))))))))))...)))))... ( -13.23) >consensus U__AAGCAGUACGCCAUUUCAUAGGUAAUAUUGAUCAAACUCCGCCUUUU__UUUUACUCAUAUCAGUAUACACACAAAUACAU____ACAUACACACAUGUGUGUAUUUCAGAAAUAUA ...............(((((..((((.................)))).....................(((((((((......................)))))))))....)))))... (-11.56 = -11.04 + -0.52)

| Location | 20,722,388 – 20,722,502 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.91 |
| Mean single sequence MFE | -21.24 |
| Consensus MFE | -15.07 |
| Energy contribution | -15.51 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20722388 114 + 20766785 UCCGCCUAUU--UUUUACUCAUAACAGUAUACACCCAAAUACAU----ACAUACACACAUGUGUGUAUUUCAGAAAUAUAAAUAGAUUUCCCAACGCUAAAUAAUGAGUUAAAGGGUAAA ...((((.((--(...((((((..............((((((((----((((......))))))))))))..(((((........))))).............)))))).)))))))... ( -22.50) >DroSec_CAF1 155734 114 + 1 UCCGCCUUUU--UCUUACUCAUAUCAGUAUACACACAAAUACAU----ACAUACACACAUGUGUGUAUUUCAGAAAUAUAAAUAGAUUUCCCAACGCUAAAUAAAGAGUUAAAGGGUAAA ...(((((((--....((((................((((((((----((((......))))))))))))..(((((........)))))...............)))).)))))))... ( -23.90) >DroSim_CAF1 160812 118 + 1 UCCGCCUUUU--UCUUACUCAUAUCAGUAUACACACAAAUACAUAUGUACAUACACACAUGUGUGUAUUUCAGAAAUAUAAAUAGAUUUCCCAACGCUAAAUAAAGAGUUAAAGGGUAAA ...(((((((--....((((..................((((((((((........))))))))))......(((((........)))))...............)))).)))))))... ( -22.50) >DroEre_CAF1 196865 109 + 1 UCCGCCUUUUUAUUUGACUCAUA---CAGUGCAUACAAAUACAU----UC----AUACAUGUGUGUAUUUCAGAAAUAUAAAUAAAUUUCCCAGCGCUAAAUAAAGAGUUAAAGAGCAAA ...((.......((((((((...---.(((((....((((((((----.(----(....)).))))))))..(((((........)))))...))))).......))))))))..))... ( -20.30) >DroYak_CAF1 162051 112 + 1 UCCGCCUUCCUUUUUUACUCAUAUCACAGUACGCACAAAUACAA----AC----AUACAUGUGUGUAUUUUAGAAAUAUAAAUCAAUUCCCCAACGCUAAAUAAAGAGUUAAAGAGCAAA .........((((((.((((.......((((((((((..((...----..----.))..)))))))))).((....))...........................)))).)))))).... ( -17.00) >consensus UCCGCCUUUU__UUUUACUCAUAUCAGUAUACACACAAAUACAU____ACAUACACACAUGUGUGUAUUUCAGAAAUAUAAAUAGAUUUCCCAACGCUAAAUAAAGAGUUAAAGGGUAAA ...(((((((......((((........(((((((((......................)))))))))....(((((........)))))...............)))).)))))))... (-15.07 = -15.51 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:38 2006