
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,714,261 – 20,714,420 |
| Length | 159 |
| Max. P | 0.999948 |

| Location | 20,714,261 – 20,714,380 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.66 |
| Mean single sequence MFE | -37.16 |
| Consensus MFE | -35.64 |
| Energy contribution | -36.24 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.47 |
| SVM RNA-class probability | 0.999904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
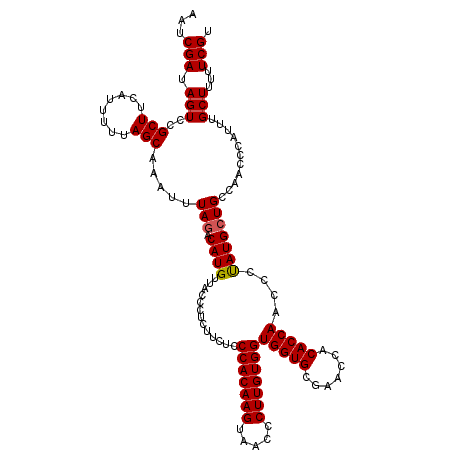
>2R_DroMel_CAF1 20714261 119 + 20766785 UGCUGCUGCUUUUUCUGUCAAA-AAAUAAGGGGAAAAGCAACGAAAAAGCAAAUGGGUUGGCAGCAUAGGGUUGGUGUGGUUCGCACCACCACAAGGAUUACUUGUGGGAGAAGAGGGUA ((((((((((((((((.(....-.....).)))))))))........(((......))))))))))......(((((((...)))))))(((((((.....)))))))............ ( -39.10) >DroSec_CAF1 147851 120 + 1 UGCUGCUGCUUUUUCUGUCAAAAAAAUAAGGGGAAAAGCAACGAAAAAGCAAAUGGGUUGGCAGCAUAGGGUUGGUGUGGUUCGCACCACCACAAGGGUUACUUGUGGGAGAAGAGGGUA ((((((((((((((((.(..........).)))))))))........(((......))))))))))......(((((((...)))))))(((((((.....)))))))............ ( -39.00) >DroSim_CAF1 152920 120 + 1 UGCUGCUGCUUUUUCUGUCAAAAAAAUAAGGGGAAAAGCAACGAAAAAGCAAAUGGGUUGGCAGCAUAGGGUUGGUGUGGUUCGCACCACCACAAGGGUUACUUGUGGGAGAAGAGGGUA ((((((((((((((((.(..........).)))))))))........(((......))))))))))......(((((((...)))))))(((((((.....)))))))............ ( -39.00) >DroEre_CAF1 188982 119 + 1 UGCUGCUGCUUUUUCUGUCAAA-AAAUAAGGGGAAAAGCAACGAAAAAGCAAAUGGGUUGACAGCAUGGGGUUGGUGUGGUUCGCACCACCACAAGGGUUACUUGUGGGAGAAGAGGGUA (((((.((((((((((.(....-.....).)))))))))).......(((......)))..)))))......(((((((...)))))))(((((((.....)))))))............ ( -34.90) >DroYak_CAF1 153902 118 + 1 UGCUGCUGCUU-UUCUGUCAAA-AAAUAAGGGGAAAAGCAACGAAAAAGCAAAUGGGUUGACAGCAUAGGGUUGGUGUGGUUUGCACCACCACAAGGGUUACUUGUGGGAGAAGAGGGUA (((((.(((((-((((.(....-......).))))))))).......(((......)))..)))))......((((((.....))))))(((((((.....)))))))............ ( -33.80) >consensus UGCUGCUGCUUUUUCUGUCAAA_AAAUAAGGGGAAAAGCAACGAAAAAGCAAAUGGGUUGGCAGCAUAGGGUUGGUGUGGUUCGCACCACCACAAGGGUUACUUGUGGGAGAAGAGGGUA ((((((((((((((((.(..........).)))))))))........(((......))))))))))......((((((.....))))))(((((((.....)))))))............ (-35.64 = -36.24 + 0.60)



| Location | 20,714,261 – 20,714,380 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.66 |
| Mean single sequence MFE | -30.40 |
| Consensus MFE | -30.27 |
| Energy contribution | -30.23 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.03 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.77 |
| SVM RNA-class probability | 0.999948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20714261 119 - 20766785 UACCCUCUUCUCCCACAAGUAAUCCUUGUGGUGGUGCGAACCACACCAACCCUAUGCUGCCAACCCAUUUGCUUUUUCGUUGCUUUUCCCCUUAUUU-UUUGACAGAAAAAGCAGCAGCA ............(((((((.....)))))))(((((.......)))))......((((((.........((((((((((((................-...))).))))))))))))))) ( -31.21) >DroSec_CAF1 147851 120 - 1 UACCCUCUUCUCCCACAAGUAACCCUUGUGGUGGUGCGAACCACACCAACCCUAUGCUGCCAACCCAUUUGCUUUUUCGUUGCUUUUCCCCUUAUUUUUUUGACAGAAAAAGCAGCAGCA ............(((((((.....)))))))(((((.......)))))......((((((.........((((((((((((....................))).))))))))))))))) ( -31.15) >DroSim_CAF1 152920 120 - 1 UACCCUCUUCUCCCACAAGUAACCCUUGUGGUGGUGCGAACCACACCAACCCUAUGCUGCCAACCCAUUUGCUUUUUCGUUGCUUUUCCCCUUAUUUUUUUGACAGAAAAAGCAGCAGCA ............(((((((.....)))))))(((((.......)))))......((((((.........((((((((((((....................))).))))))))))))))) ( -31.15) >DroEre_CAF1 188982 119 - 1 UACCCUCUUCUCCCACAAGUAACCCUUGUGGUGGUGCGAACCACACCAACCCCAUGCUGUCAACCCAUUUGCUUUUUCGUUGCUUUUCCCCUUAUUU-UUUGACAGAAAAAGCAGCAGCA ............(((((((.....)))))))(((((.......)))))......((((((.........((((((((((((................-...))).))))))))))))))) ( -29.81) >DroYak_CAF1 153902 118 - 1 UACCCUCUUCUCCCACAAGUAACCCUUGUGGUGGUGCAAACCACACCAACCCUAUGCUGUCAACCCAUUUGCUUUUUCGUUGCUUUUCCCCUUAUUU-UUUGACAGAA-AAGCAGCAGCA ............(((((((.....)))))))(((((.......)))))......(((((.(((.....))))......((((((((((...(((...-..)))..)))-))))))))))) ( -28.70) >consensus UACCCUCUUCUCCCACAAGUAACCCUUGUGGUGGUGCGAACCACACCAACCCUAUGCUGCCAACCCAUUUGCUUUUUCGUUGCUUUUCCCCUUAUUU_UUUGACAGAAAAAGCAGCAGCA ............(((((((.....)))))))(((((.......)))))......((((((.........((((((((((((....................))).))))))))))))))) (-30.27 = -30.23 + -0.04)

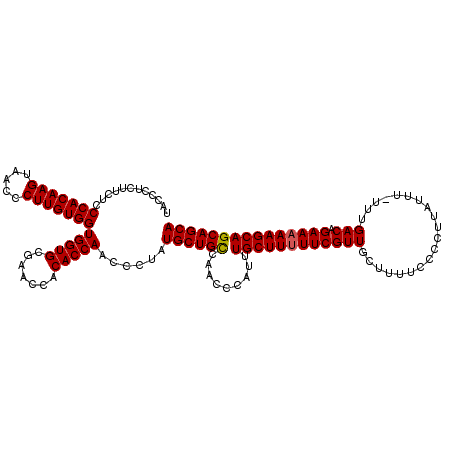
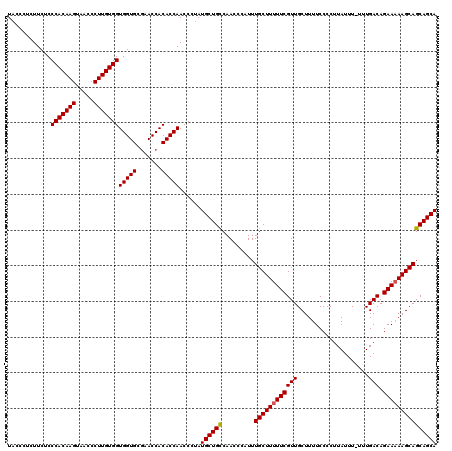
| Location | 20,714,300 – 20,714,420 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -30.24 |
| Energy contribution | -29.68 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20714300 120 + 20766785 ACGAAAAAGCAAAUGGGUUGGCAGCAUAGGGUUGGUGUGGUUCGCACCACCACAAGGAUUACUUGUGGGAGAAGAGGGUAACAUGUCUAAAUUUGCUAAAAAUGAAGCGGACUAUCGAUU .(((....((..((...(((((((..((((..(((((((...)))))))(((((((.....))))))).................))))...)))))))..))...))......)))... ( -31.00) >DroSec_CAF1 147891 120 + 1 ACGAAAAAGCAAAUGGGUUGGCAGCAUAGGGUUGGUGUGGUUCGCACCACCACAAGGGUUACUUGUGGGAGAAGAGGGUAACAUGUCUAAAUUUGCUAAAAAUGAAGCGGACUAUCGAUU .(((....((..((...(((((((..((((..(((((((...)))))))(((((((.....))))))).................))))...)))))))..))...))......)))... ( -31.00) >DroSim_CAF1 152960 120 + 1 ACGAAAAAGCAAAUGGGUUGGCAGCAUAGGGUUGGUGUGGUUCGCACCACCACAAGGGUUACUUGUGGGAGAAGAGGGUAACAUGUCUAAAUUUGCUAAAAAUGAAGCGAACUAUCGAUU .......(((((((.....((((.........(((((((...)))))))(((((((.....)))))))...............))))...)))))))..........(((....)))... ( -31.10) >DroEre_CAF1 189021 120 + 1 ACGAAAAAGCAAAUGGGUUGACAGCAUGGGGUUGGUGUGGUUCGCACCACCACAAGGGUUACUUGUGGGAGAAGAGGGUAACAUGUAUAAAUUUGCUAAAAAUGAAGCGGACUAUCGAUU .(((...(((((((..((.....(((((....(((((((...)))))))(((((((.....))))))).............)))))))..)))))))........((....)).)))... ( -31.60) >DroYak_CAF1 153940 120 + 1 ACGAAAAAGCAAAUGGGUUGACAGCAUAGGGUUGGUGUGGUUUGCACCACCACAAGGGUUACUUGUGGGAGAAGAGGGUAACAUGUCUAAAUUUGCUAAAAAUGAAGCGGACUAUCGAUU .(((...(((((((.....((((.........((((((.....))))))(((((((.....)))))))...............))))...)))))))........((....)).)))... ( -30.80) >consensus ACGAAAAAGCAAAUGGGUUGGCAGCAUAGGGUUGGUGUGGUUCGCACCACCACAAGGGUUACUUGUGGGAGAAGAGGGUAACAUGUCUAAAUUUGCUAAAAAUGAAGCGGACUAUCGAUU .(((................(((.((......)).)))(((((((.(..(((((((.....)))))))..).....(((((.((......))))))).........))))))).)))... (-30.24 = -29.68 + -0.56)

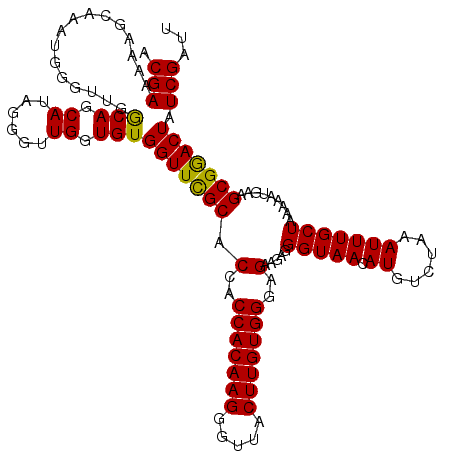
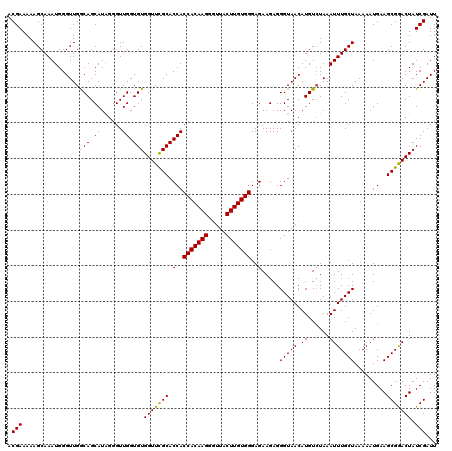
| Location | 20,714,300 – 20,714,420 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -23.24 |
| Consensus MFE | -21.66 |
| Energy contribution | -21.70 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20714300 120 - 20766785 AAUCGAUAGUCCGCUUCAUUUUUAGCAAAUUUAGACAUGUUACCCUCUUCUCCCACAAGUAAUCCUUGUGGUGGUGCGAACCACACCAACCCUAUGCUGCCAACCCAUUUGCUUUUUCGU ...(((.(((..(((........))).....(((.((((.............(((((((.....)))))))(((((.......)))))....)))))))...........)))...))). ( -22.40) >DroSec_CAF1 147891 120 - 1 AAUCGAUAGUCCGCUUCAUUUUUAGCAAAUUUAGACAUGUUACCCUCUUCUCCCACAAGUAACCCUUGUGGUGGUGCGAACCACACCAACCCUAUGCUGCCAACCCAUUUGCUUUUUCGU ...(((.(((..(((........))).....(((.((((.............(((((((.....)))))))(((((.......)))))....)))))))...........)))...))). ( -22.40) >DroSim_CAF1 152960 120 - 1 AAUCGAUAGUUCGCUUCAUUUUUAGCAAAUUUAGACAUGUUACCCUCUUCUCCCACAAGUAACCCUUGUGGUGGUGCGAACCACACCAACCCUAUGCUGCCAACCCAUUUGCUUUUUCGU ...(((.(((..(((........))).....(((.((((.............(((((((.....)))))))(((((.......)))))....)))))))...........)))...))). ( -22.40) >DroEre_CAF1 189021 120 - 1 AAUCGAUAGUCCGCUUCAUUUUUAGCAAAUUUAUACAUGUUACCCUCUUCUCCCACAAGUAACCCUUGUGGUGGUGCGAACCACACCAACCCCAUGCUGUCAACCCAUUUGCUUUUUCGU ....(((((...(((........))).........((((.............(((((((.....)))))))(((((.......)))))....)))))))))................... ( -24.70) >DroYak_CAF1 153940 120 - 1 AAUCGAUAGUCCGCUUCAUUUUUAGCAAAUUUAGACAUGUUACCCUCUUCUCCCACAAGUAACCCUUGUGGUGGUGCAAACCACACCAACCCUAUGCUGUCAACCCAUUUGCUUUUUCGU ...(((.(((..(((........))).......((((.((............(((((((.....)))))))(((((.......))))).......)))))).........)))...))). ( -24.30) >consensus AAUCGAUAGUCCGCUUCAUUUUUAGCAAAUUUAGACAUGUUACCCUCUUCUCCCACAAGUAACCCUUGUGGUGGUGCGAACCACACCAACCCUAUGCUGCCAACCCAUUUGCUUUUUCGU ...(((.(((..(((........))).....(((.((((.............(((((((.....)))))))(((((.......)))))....)))))))...........)))...))). (-21.66 = -21.70 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:32 2006