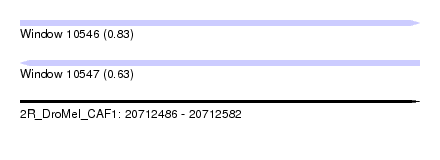
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,712,486 – 20,712,582 |
| Length | 96 |
| Max. P | 0.828209 |

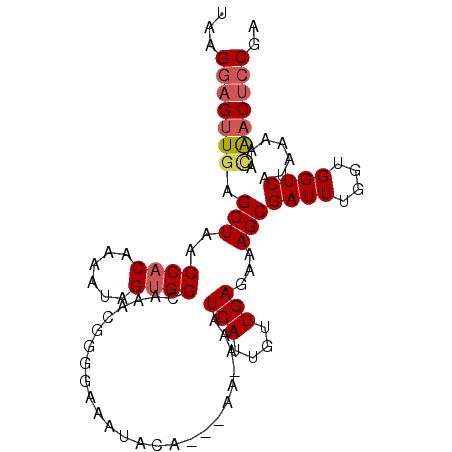
| Location | 20,712,486 – 20,712,582 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 87.84 |
| Mean single sequence MFE | -22.08 |
| Consensus MFE | -14.65 |
| Energy contribution | -15.53 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20712486 96 + 20766785 UAAGGAGUUGAGCUAACCACAAAAUAGUGGCAAACGGUGAAAUACA---AAAAAAUCAUUGUUGAGAAAGCGAUUUGGUGGUCAUAAAAAGAACUACGA ....((((((..((..((((......))))..((((((((......---......)))))))).))....)))))).((((((.......).))))).. ( -16.90) >DroSec_CAF1 146061 95 + 1 UAAGGAGUUGAGCUAACCACAAAAUAGCGGCAAACGGUGAAAUACA---AA-AAAUCAUUGUUGAGAAAGCGAUUUGGUGGUCAUAAAAACAACUCCGA ...(((((((.....(((((.((((.((..(.((((((((......---..-...))))))))..)...)).)))).)))))........))))))).. ( -26.52) >DroEre_CAF1 187034 98 + 1 UAAGGAGUUGAGCUAACCACAAAAUAGUGGCAA-CGGGAAAAUACAACAAAAAAAUCAUUGUUGAGAAAGCGAUUUGGUGGUCAUAAAAAUGCCGCCGA ...........(((..((((......))))...-..........((((((........))))))....)))...(((((((.(((....)))))))))) ( -23.00) >DroYak_CAF1 151979 86 + 1 UAAGGAGUUGAGCUAACCACAAAAUAGUGGCAAACGGGG-------------AAAUCAUUGUUGAGAAAGCGAUUUGGUGGUCAUAAAAACGACUCCGA ...(((((((.(((..((((......))))..(((((.(-------------....).))))).....)))((((....)))).......))))))).. ( -21.90) >consensus UAAGGAGUUGAGCUAACCACAAAAUAGUGGCAAACGGGGAAAUACA___AA_AAAUCAUUGUUGAGAAAGCGAUUUGGUGGUCAUAAAAACAACUCCGA ...(((((((.(((..((((......)))).........................(((....)))...)))((((....)))).......))))))).. (-14.65 = -15.53 + 0.88)

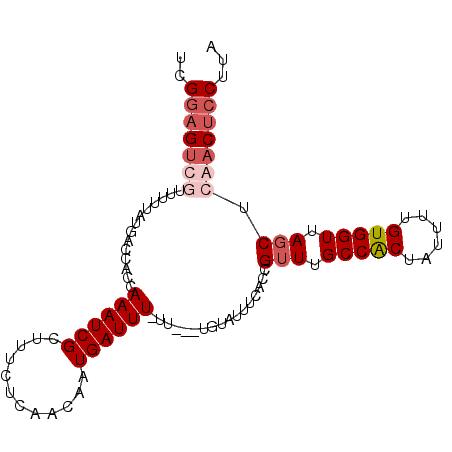
| Location | 20,712,486 – 20,712,582 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 87.84 |
| Mean single sequence MFE | -18.48 |
| Consensus MFE | -11.74 |
| Energy contribution | -13.55 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20712486 96 - 20766785 UCGUAGUUCUUUUUAUGACCACCAAAUCGCUUUCUCAACAAUGAUUUUUU---UGUAUUUCACCGUUUGCCACUAUUUUGUGGUUAGCUCAACUCCUUA ((((((......))))))...................((((........)---)))........(((.(((((......))))).)))........... ( -13.60) >DroSec_CAF1 146061 95 - 1 UCGGAGUUGUUUUUAUGACCACCAAAUCGCUUUCUCAACAAUGAUUU-UU---UGUAUUUCACCGUUUGCCGCUAUUUUGUGGUUAGCUCAACUCCUUA ..(((((((......(((((((.((((.((......(((..(((..(-..---...)..)))..)))....)).)))).)))))))...)))))))... ( -23.80) >DroEre_CAF1 187034 98 - 1 UCGGCGGCAUUUUUAUGACCACCAAAUCGCUUUCUCAACAAUGAUUUUUUUGUUGUAUUUUCCCG-UUGCCACUAUUUUGUGGUUAGCUCAACUCCUUA ..((.(((((....))).)).)).....(((....((((((........))))))..........-..(((((......))))).)))........... ( -20.10) >DroYak_CAF1 151979 86 - 1 UCGGAGUCGUUUUUAUGACCACCAAAUCGCUUUCUCAACAAUGAUUU-------------CCCCGUUUGCCACUAUUUUGUGGUUAGCUCAACUCCUUA ..(((((................((((((............))))))-------------....(((.(((((......))))).)))...)))))... ( -16.40) >consensus UCGGAGUCGUUUUUAUGACCACCAAAUCGCUUUCUCAACAAUGAUUU_UU___UGUAUUUCACCGUUUGCCACUAUUUUGUGGUUAGCUCAACUCCUUA ..(((((((..............((((((............)))))).................(((.(((((......))))).))).)))))))... (-11.74 = -13.55 + 1.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:27 2006