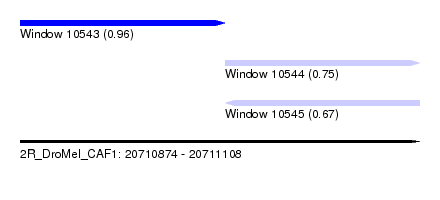
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,710,874 – 20,711,108 |
| Length | 234 |
| Max. P | 0.959565 |

| Location | 20,710,874 – 20,710,994 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.66 |
| Mean single sequence MFE | -29.26 |
| Consensus MFE | -25.24 |
| Energy contribution | -25.64 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20710874 120 + 20766785 AACGUAUACGAAGACGCAGUUCCAAGUGCGUCCUGUAGGUGUUCCCCUGGAAGACAUCUGCCAAGCCGACUUCCUUUGGUGUAAACCAAUUUUCGUAUCAAUAAAAUUAAAAUCUACUAU ...(.(((((((((((((.(....).))))))..(((((((((((...)))..))))))))..............(((((....)))))..))))))))..................... ( -29.40) >DroSec_CAF1 144411 119 + 1 AACGUAUACGAAGACGCAGUCCCAAGUGAGUCCUGUAGGUGUUC-CCUGCAAGACAUCUGCCAAGCCGACUUCCUUUGGUGUAAACCAAUUUUCGUAUCAAUACAAUUAAAAUCCACUAU ...(.(((((((((.((((((......))(((.((((((.....-)))))).)))..))))..............(((((....)))))))))))))))..................... ( -28.00) >DroSim_CAF1 149402 119 + 1 AACGUAUACGAAGACGCAGUCCCAAGUGCGUCCUGUAGGUGUUC-CCUGCAAGACAUCUGCCAAGCCGACUUCCUUUGGUGUAAACCAAUUUUCGUAUCAAUACAAUUAAAAUCCACUAU ...(.(((((((((((((.(....).))))))..(((((((((.-.......)))))))))..............(((((....)))))..))))))))..................... ( -29.00) >DroEre_CAF1 185591 120 + 1 AACGUAUACGAAGACGCAGUCCCACGUGCGUCCUGUAGGUGUUUCCCUGCAAGACAGCUGCCAAGCCGACUUCCUUUGGUGUAAACCAAUUUUCGUAUCAAUAAAAUUGAAAUCCAUUAA ...(.(((((((((.(.((((........(((.((((((......)))))).))).(((....))).)))).)..(((((....)))))))))))))))..................... ( -30.70) >DroYak_CAF1 150486 120 + 1 AACGUAUACGAAUACGCAGUCCCACGUGCGUCCUGUAGGUGUUUCCCUGCAAGACAGCUGCCCUGCCGACUUCCUUUGGUGUAAACCAAUUUUCGUAUCAAUAAAAUUGAAAUCCAUUAA ...(.(((((((...((((...((.((..(((.((((((......)))))).))).))))..)))).........(((((....)))))..))))))))..................... ( -29.20) >consensus AACGUAUACGAAGACGCAGUCCCAAGUGCGUCCUGUAGGUGUUCCCCUGCAAGACAUCUGCCAAGCCGACUUCCUUUGGUGUAAACCAAUUUUCGUAUCAAUAAAAUUAAAAUCCACUAU ...(.(((((((((.((((...(....).(((.((((((......)))))).)))..))))..............(((((....)))))))))))))))..................... (-25.24 = -25.64 + 0.40)

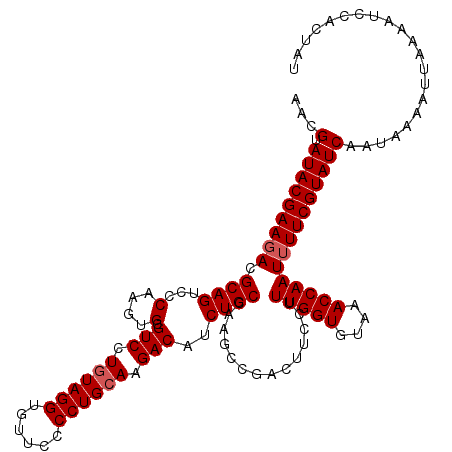
| Location | 20,710,994 – 20,711,108 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.62 |
| Mean single sequence MFE | -32.34 |
| Consensus MFE | -25.16 |
| Energy contribution | -25.00 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20710994 114 + 20766785 AGGUCUUCGUCU-UCGAACCAAAAUCGAAAUUUGGCUGUUUU----UUUAAGGGGCUCGAUAGCCUUUGUCAAAGGGCCACUAAUUGUGCUGCUAAAGAACGAA-CGAAAGAGAAAACUG .(((.(((.(((-(((..(((((.......))))).((((((----((..((.(((.((((.((((((....)))))).....)))).))).))))))))))..-)).))))))).))). ( -29.50) >DroSec_CAF1 144530 115 + 1 AGGUCUUCGUCUUUCGAAGCAAAAUCGAAAUUUGGCUGUUUU----UUUAAGGGGCUCGAUAGCCUUUGUCAAAGGGGCACUAAUUGUGCUGCUAAAGAACGAA-UGAAAGAGAAAACUG .(((.(((.(((((((.(((.((((....)))).)))(((((----((..(((((((....))))))).....((.(((((.....))))).)))))))))...-)))))))))).))). ( -33.60) >DroSim_CAF1 149521 115 + 1 AGGUCUUCGUCUUUCGAAGCAAAAUCGAAAUUUGGCUGUUUU----UUUAAGGGGCUCGAUAGCCUUUGUCAAAGGGGCACUAAUUGUGCUGCUAAAGAACGAA-UGAAAGAGAAAACUG .(((.(((.(((((((.(((.((((....)))).)))(((((----((..(((((((....))))))).....((.(((((.....))))).)))))))))...-)))))))))).))). ( -33.60) >DroEre_CAF1 185711 114 + 1 AGGUCUUCGGCUUUCGAACCGAAAUCGAAAUUUGGCUGUUUUUUUUUUUAAGGGG------AGCCUUUGUCAAAAGGUCACUAAUUGUGCUGCCGAAGAACGAAAGGAAAGCGAAAACUG ...((((((((((((((.......))))))...((((.(((((......))))).------))))..........((.(((.....))))))))))))).(....).............. ( -29.80) >DroYak_CAF1 150606 116 + 1 AGGUCUUCGGCUUUCGAACCGAAAUCGAAAUUUGGCUGUUUU----UCUAAGGGGCUCGAUAGCCUUUGUCAAAAGGCCACUAAUUGUGCUGCUGAAGAACGAAAGGAAAGCGAAAACUG .(((.((((.(((((...(((((.......))))).((((((----((..((.(((.((((.((((((....)))))).....)))).))).))))))))))....))))))))).))). ( -35.20) >consensus AGGUCUUCGUCUUUCGAACCAAAAUCGAAAUUUGGCUGUUUU____UUUAAGGGGCUCGAUAGCCUUUGUCAAAGGGCCACUAAUUGUGCUGCUAAAGAACGAA_GGAAAGAGAAAACUG .(((.(((.(((((((........(((...((((((..............((((((......)))))).......((.(((.....)))))))))))...)))..)))))))))).))). (-25.16 = -25.00 + -0.16)

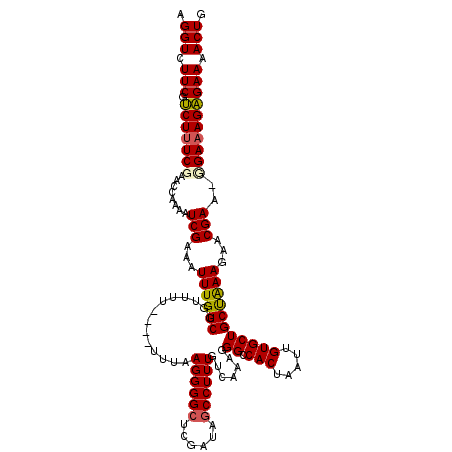
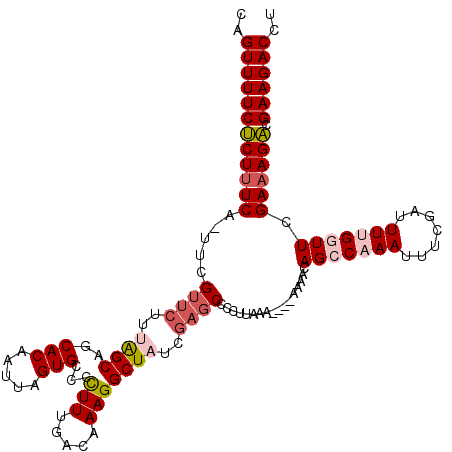
| Location | 20,710,994 – 20,711,108 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.62 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -19.80 |
| Energy contribution | -20.92 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20710994 114 - 20766785 CAGUUUUCUCUUUCG-UUCGUUCUUUAGCAGCACAAUUAGUGGCCCUUUGACAAAGGCUAUCGAGCCCCUUAAA----AAAACAGCCAAAUUUCGAUUUUGGUUCGA-AGACGAAGACCU ..((((((((((.((-...((((..((((..(((.....)))...(((.....)))))))..))))........----.....(((((((.......))))))))))-))).)))))).. ( -25.10) >DroSec_CAF1 144530 115 - 1 CAGUUUUCUCUUUCA-UUCGUUCUUUAGCAGCACAAUUAGUGCCCCUUUGACAAAGGCUAUCGAGCCCCUUAAA----AAAACAGCCAAAUUUCGAUUUUGCUUCGAAAGACGAAGACCU ..((((((((((((.-...((((..((((.((((.....))))..(((.....)))))))..))))........----.....(((.((((....)))).)))..)))))).)))))).. ( -28.30) >DroSim_CAF1 149521 115 - 1 CAGUUUUCUCUUUCA-UUCGUUCUUUAGCAGCACAAUUAGUGCCCCUUUGACAAAGGCUAUCGAGCCCCUUAAA----AAAACAGCCAAAUUUCGAUUUUGCUUCGAAAGACGAAGACCU ..((((((((((((.-...((((..((((.((((.....))))..(((.....)))))))..))))........----.....(((.((((....)))).)))..)))))).)))))).. ( -28.30) >DroEre_CAF1 185711 114 - 1 CAGUUUUCGCUUUCCUUUCGUUCUUCGGCAGCACAAUUAGUGACCUUUUGACAAAGGCU------CCCCUUAAAAAAAAAAACAGCCAAAUUUCGAUUUCGGUUCGAAAGCCGAAGACCU ...................(.((((((((..(((.....))).............((((------..................))))...((((((.......)))))))))))))).). ( -24.17) >DroYak_CAF1 150606 116 - 1 CAGUUUUCGCUUUCCUUUCGUUCUUCAGCAGCACAAUUAGUGGCCUUUUGACAAAGGCUAUCGAGCCCCUUAGA----AAAACAGCCAAAUUUCGAUUUCGGUUCGAAAGCCGAAGACCU ..((((((((((((.........................(((((((((....))))))))).(((((.....((----((..........))))......)))))))))).))))))).. ( -29.50) >consensus CAGUUUUCUCUUUCA_UUCGUUCUUUAGCAGCACAAUUAGUGCCCCUUUGACAAAGGCUAUCGAGCCCCUUAAA____AAAACAGCCAAAUUUCGAUUUUGGUUCGAAAGACGAAGACCU ..((((((((((((.....((((..((((..(((.....)))...(((.....)))))))..)))).................(((((((.......))))))).)))))).)))))).. (-19.80 = -20.92 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:26 2006