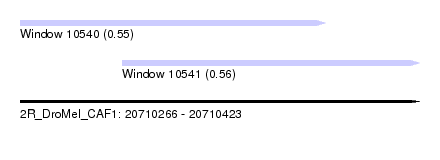
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,710,266 – 20,710,423 |
| Length | 157 |
| Max. P | 0.559429 |

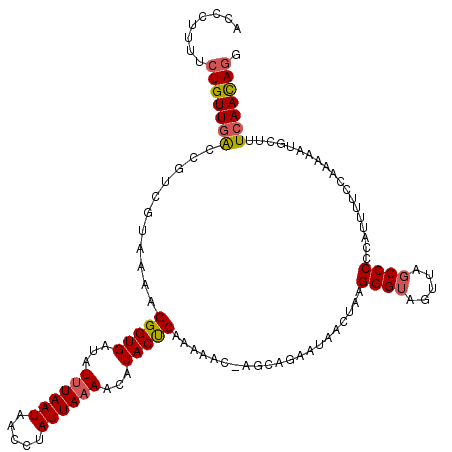
| Location | 20,710,266 – 20,710,386 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.73 |
| Mean single sequence MFE | -18.34 |
| Consensus MFE | -13.36 |
| Energy contribution | -13.36 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20710266 120 + 20766785 CUGUGUCAUCAGCAUCAGGCACAAACAAUCAAAUAAAGCAACCCUUUUCUGUUGACCGUCGUAAAAGGGUGAUAUUUAAUAACCUAUUAAAACAUACUCAAAAACAAGCAGAAUAACUAA .((((((..........))))))..............((.((((((((.((........)).))))))))....(((((((...)))))))................))........... ( -18.90) >DroSec_CAF1 143829 110 + 1 CGGUGUCAUCAACAUCAGACAC----AAUCAAAUAAAGCAACCCUUUUCUGUUGACCGUCGUAAAAGGGUGAUAUUUAAUAACCCAUUAAAACAUACUCA------AGGAGAAUAACUUA ..(((((..........)))))----.......((((...((((((((.((........)).))))))))....))))..................(((.------..)))......... ( -17.00) >DroSim_CAF1 148820 110 + 1 CGGUGUCAUCAACAUCAGACAC----AAUCAAAUAAAGCAACCCUUUUCUGUUGACCGUCGUAAAAGGGUGAUAUUUAAUAACCCAUUAAAACAUACUCA------AGGAGAAUAACUAA ..(((((..........)))))----.......((((...((((((((.((........)).))))))))....))))..................(((.------..)))......... ( -17.00) >DroEre_CAF1 184980 120 + 1 CGGUGUCAUCAGCAUCAAACACAAACAAUCAAAUAAAGCAACCCUUUUCUGUUUGCCUUCGUAAGAGGGUGAUAUUUAAUAACCUAUUAAAACAUACUCAAAAACCACCAGAAUAACUAA .(((((.....)))))..............................(((((.(..(((((....)))))..)..(((((((...))))))).................)))))....... ( -20.20) >DroYak_CAF1 149860 118 + 1 CGGUGUCAUCAGCAUCAAACACAAACAAUCAAACAAAGCAACCCUUUUCUGUUGGCCUUCGUAAAAGGGUGAUAUAUAAUAAACUAUUA-AACAUACCCAAAAACCAGCAGAAU-ACUAA .(((((.....)))))..............................((((((((((....).....(((((.....(((((...)))))-....))))).....))))))))).-..... ( -18.60) >consensus CGGUGUCAUCAGCAUCAGACACAAACAAUCAAAUAAAGCAACCCUUUUCUGUUGACCGUCGUAAAAGGGUGAUAUUUAAUAACCUAUUAAAACAUACUCAAAAAC_AGCAGAAUAACUAA .(((((.....)))))............((..........((((((((.((........)).))))))))....((((((.....))))))...................))........ (-13.36 = -13.36 + 0.00)

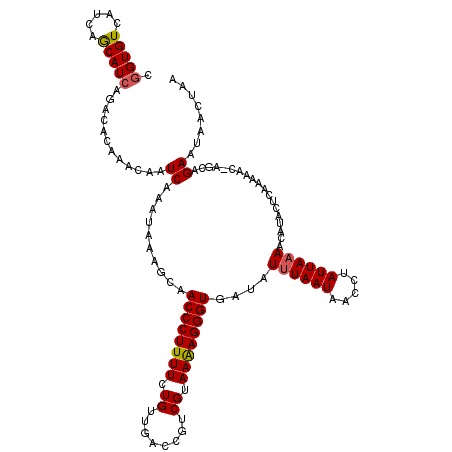
| Location | 20,710,306 – 20,710,423 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.02 |
| Mean single sequence MFE | -24.66 |
| Consensus MFE | -16.16 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20710306 117 + 20766785 ACCCUUUUCUGUUGACCGUCGUAAAAGGGUGAUAUUUAAUAACCUAUUAAAACAUACUCAAAAACAAGCAGAAUAACUAAGGGUGGU---CCCCCAUUUUCCAAAAAUGCUUUCAACAAG .........((((((...........(((((...(((((((...)))))))...)))))......(((((.........(((((((.---...))))))).......))))))))))).. ( -19.09) >DroSec_CAF1 143865 114 + 1 ACCCUUUUCUGUUGACCGUCGUAAAAGGGUGAUAUUUAAUAACCCAUUAAAACAUACUCA------AGGAGAAUAACUUAGGGUAGUUAGCCCCCAUUUUCCAAAAAUGAUUUCAACAGG ........(((((((..(((((....((((...........))))...............------.(((((((......((((.....))))..)))))))....))))).))))))). ( -28.00) >DroSim_CAF1 148856 114 + 1 ACCCUUUUCUGUUGACCGUCGUAAAAGGGUGAUAUUUAAUAACCCAUUAAAACAUACUCA------AGGAGAAUAACUAAGGGUAGUUAGCCCCCAUUUUCCAAACAUGAUUUCAACAGG ........(((((((..(((((....((((...........))))...............------.(((((((......((((.....))))..)))))))....))))).))))))). ( -28.00) >DroEre_CAF1 185020 120 + 1 ACCCUUUUCUGUUUGCCUUCGUAAGAGGGUGAUAUUUAAUAACCUAUUAAAACAUACUCAAAAACCACCAGAAUAACUAAGGGUAGUUAGCCCCUAUUUUACAAAAAUGCUUUCAACAGG (((((((((((.(..(((((....)))))..)..(((((((...))))))).................))))).....)))))).(..(((.................)))..)...... ( -23.63) >DroYak_CAF1 149900 118 + 1 ACCCUUUUCUGUUGGCCUUCGUAAAAGGGUGAUAUAUAAUAAACUAUUA-AACAUACCCAAAAACCAGCAGAAU-ACUAAGGGUAGUUAGCCCCUAUCUUAGAAAUAUGCUUUCAAUAGG ((((((((((((((((....).....(((((.....(((((...)))))-....))))).....))))))))).-...))))))........(((((....((((.....)))).))))) ( -24.60) >consensus ACCCUUUUCUGUUGACCGUCGUAAAAGGGUGAUAUUUAAUAACCUAUUAAAACAUACUCAAAAAC_AGCAGAAUAACUAAGGGUAGUUAGCCCCCAUUUUCCAAAAAUGCUUUCAACAGG ........(((((((...........(((((...((((((.....))))))...))))).....................((((.....))))...................))))))). (-16.16 = -16.40 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:22 2006