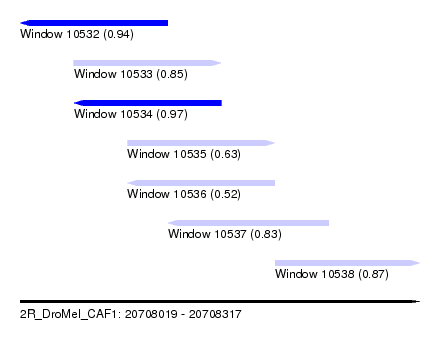
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,708,019 – 20,708,317 |
| Length | 298 |
| Max. P | 0.973511 |

| Location | 20,708,019 – 20,708,129 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.36 |
| Mean single sequence MFE | -23.72 |
| Consensus MFE | -19.20 |
| Energy contribution | -20.36 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20708019 110 - 20766785 UUGCAAAUUGAUACGAAA----------AAAAUGCAAUGACAGAAAUACAAAGGAAACAGGAACUGUGAUGGGUUGGAAAAGGGGGAGAUGGCCCACGGCUGAUUAUAGGGUUGCGUUGA ..................----------..((((((((..............(....).....(((((((.(((((......(((.......))).))))).))))))).)))))))).. ( -25.90) >DroSec_CAF1 141764 114 - 1 UUGCAAAUUGAUACGAAAAA------AAAAAAUGCAAUGACAGAAAUACAAAGGAAACAGGGACUGUGAUGGGUUGGAAAAGGGAGAGAUGGCCCACGGCUGAUUAUAGGGUUGCGUUGA ....................------....((((((((..............(....).....(((((((.(((((.....(((........))).))))).))))))).)))))))).. ( -25.70) >DroSim_CAF1 146763 112 - 1 UUGCAAAUUGAUACGAAAA--------AAAAAUGCAAUGACAGAAAUACAAAGGAAACAGGGACUGUGAUGGGUUGGAAAAGGGAGAGAUGGCCCACGGCUGAUUAUAGGGUUGCGUUGA ...................--------...((((((((..............(....).....(((((((.(((((.....(((........))).))))).))))))).)))))))).. ( -25.70) >DroEre_CAF1 182405 120 - 1 UUGCAAAUUGAUACAAAAAAAAAAAAAAAAAAUGCAAUGACAGAAAUACAAAGGAAACAGGGGUUUUGUUGGGUUGGUAAAGGGAGAGAUGGCCCACGGCUGAUUAUAGGGUUGCGUCGA ...............................((((((((((((((...(...(....)..)..))))))).(((..((...(((........)))...))..))).....)))))))... ( -22.30) >DroYak_CAF1 147791 111 - 1 UUGCAAAUUGAUACAAAA---------AAAAAUGCAAUGACAGAAAUACAAAAGAAACAGGGGUUUUGUUGGGUUGGUAAAGGGAGAGAUGGCCCACGGCUGAUUAUAGGGUUGCGUCGA ..................---------....((((((((((((((...(..........)...))))))).(((..((...(((........)))...))..))).....)))))))... ( -19.00) >consensus UUGCAAAUUGAUACGAAAA________AAAAAUGCAAUGACAGAAAUACAAAGGAAACAGGGACUGUGAUGGGUUGGAAAAGGGAGAGAUGGCCCACGGCUGAUUAUAGGGUUGCGUUGA ..............................((((((((..............(....).....(((((((.(((((.....(((........))).))))).))))))).)))))))).. (-19.20 = -20.36 + 1.16)

| Location | 20,708,059 – 20,708,169 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.20 |
| Mean single sequence MFE | -11.99 |
| Consensus MFE | -10.63 |
| Energy contribution | -10.13 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20708059 110 + 20766785 UUUCCAACCCAUCACAGUUCCUGUUUCCUUUGUAUUUCUGUCAUUGCAUUUU----------UUUCGUAUCAAUUUGCAAUUUUUUCGCUUAUUUUCGAGUUGUUUUUCACCAACUCGCC .............((((...))))..................((((((....----------.............))))))...............(((((((........))))))).. ( -12.13) >DroSim_CAF1 146803 112 + 1 UUUCCAACCCAUCACAGUCCCUGUUUCCUUUGUAUUUCUGUCAUUGCAUUUUU--------UUUUCGUAUCAAUUUGCAAUUUUUUCGCUUAUUUUCGAGUUGUUUUUCACCAACUCGCC .............((((...))))..................((((((.....--------..............))))))...............(((((((........))))))).. ( -12.01) >DroEre_CAF1 182445 120 + 1 UUACCAACCCAACAAAACCCCUGUUUCCUUUGUAUUUCUGUCAUUGCAUUUUUUUUUUUUUUUUUUGUAUCAAUUUGCAAUUUUUUCGCUCAUUUUCAAGUUGUUUUUCACCAACUUGCC ...........(((((............))))).........((((((...........................))))))...............(((((((........))))))).. ( -9.63) >DroYak_CAF1 147831 111 + 1 UUACCAACCCAACAAAACCCCUGUUUCUUUUGUAUUUCUGUCAUUGCAUUUUU---------UUUUGUAUCAAUUUGCAAUUUUUGUGCCCAUUUUCAAGUUGUUUUUCACCAACUUGCC ...........((((((..........))))))............((((....---------..(((((......))))).....)))).......(((((((........))))))).. ( -14.20) >consensus UUACCAACCCAACAAAACCCCUGUUUCCUUUGUAUUUCUGUCAUUGCAUUUUU_________UUUCGUAUCAAUUUGCAAUUUUUUCGCUCAUUUUCAAGUUGUUUUUCACCAACUCGCC ..........................................((((((...........................))))))...............(((((((........))))))).. (-10.63 = -10.13 + -0.50)

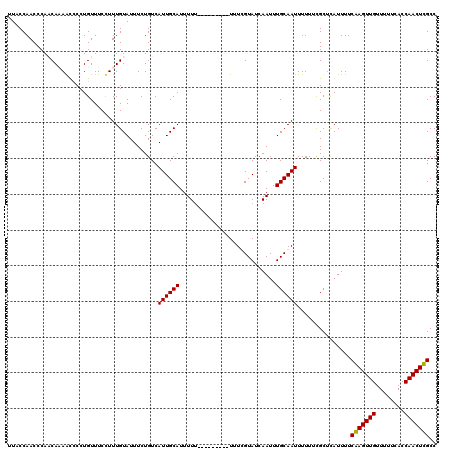
| Location | 20,708,059 – 20,708,169 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.20 |
| Mean single sequence MFE | -18.61 |
| Consensus MFE | -14.92 |
| Energy contribution | -14.98 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20708059 110 - 20766785 GGCGAGUUGGUGAAAAACAACUCGAAAAUAAGCGAAAAAAUUGCAAAUUGAUACGAAA----------AAAAUGCAAUGACAGAAAUACAAAGGAAACAGGAACUGUGAUGGGUUGGAAA ..(((((((........))))))).......((((.....))))..............----------..(((.((.(.((((.........(....).....)))).))).)))..... ( -17.84) >DroSim_CAF1 146803 112 - 1 GGCGAGUUGGUGAAAAACAACUCGAAAAUAAGCGAAAAAAUUGCAAAUUGAUACGAAAA--------AAAAAUGCAAUGACAGAAAUACAAAGGAAACAGGGACUGUGAUGGGUUGGAAA ..(((((((........))))))).......((((.....))))...............--------...(((.((.(.((((.....(...(....)..)..)))).))).)))..... ( -18.90) >DroEre_CAF1 182445 120 - 1 GGCAAGUUGGUGAAAAACAACUUGAAAAUGAGCGAAAAAAUUGCAAAUUGAUACAAAAAAAAAAAAAAAAAAUGCAAUGACAGAAAUACAAAGGAAACAGGGGUUUUGUUGGGUUGGUAA ..(((((((........)))))))...((.(((......((((((...........................))))))(((((((...(...(....)..)..)))))))..))).)).. ( -20.33) >DroYak_CAF1 147831 111 - 1 GGCAAGUUGGUGAAAAACAACUUGAAAAUGGGCACAAAAAUUGCAAAUUGAUACAAAA---------AAAAAUGCAAUGACAGAAAUACAAAAGAAACAGGGGUUUUGUUGGGUUGGUAA ..(((((((........)))))))...((.(((......((((((.............---------.....))))))(((((((...(..........)...)))))))..))).)).. ( -17.37) >consensus GGCAAGUUGGUGAAAAACAACUCGAAAAUAAGCGAAAAAAUUGCAAAUUGAUACAAAA_________AAAAAUGCAAUGACAGAAAUACAAAGGAAACAGGGACUGUGAUGGGUUGGAAA ..(((((((........)))))))...............((((((...........................))))))..(((....((((((..........))))))....))).... (-14.92 = -14.98 + 0.06)

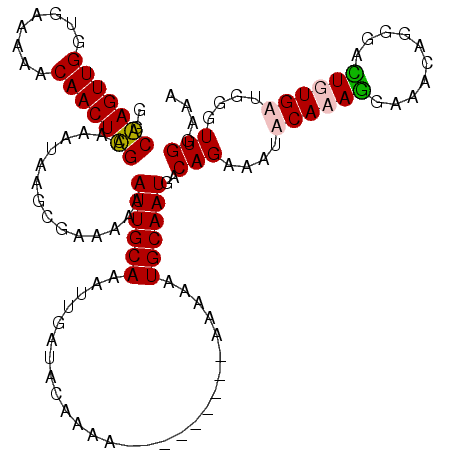
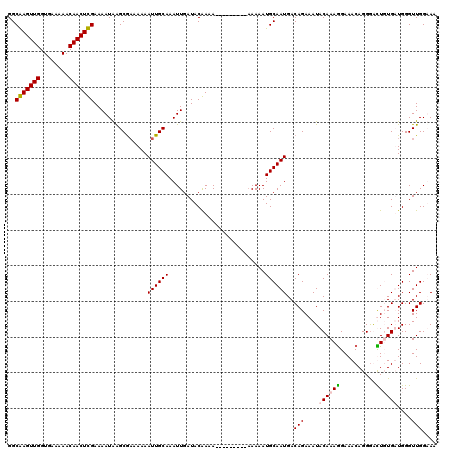
| Location | 20,708,099 – 20,708,209 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.80 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -18.39 |
| Energy contribution | -18.23 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20708099 110 + 20766785 UCAUUGCAUUUU----------UUUCGUAUCAAUUUGCAAUUUUUUCGCUUAUUUUCGAGUUGUUUUUCACCAACUCGCCGACUUGGUGGUUGUGCUAGAAGUUGAUUAUGCCCGACGAU ............----------..((((....((...((((((((............((((((........))))))(((((((....))))).)).))))))))...)).....)))). ( -22.10) >DroSec_CAF1 141844 113 + 1 UCAUUGCAUUUUUU------UUUUUCGUAUCAAUUUGCAAUUUUUUCGCUUAUUUUCGAGUUGUUUUUCACCAACUCGCCGACUUGGUGGUUGUGCUAGAAGUUGAUU-UGCCCGACGAU ..............------....((((..(((....((((((((............((((((........))))))(((((((....))))).)).))))))))..)-))....)))). ( -22.50) >DroSim_CAF1 146843 112 + 1 UCAUUGCAUUUUU--------UUUUCGUAUCAAUUUGCAAUUUUUUCGCUUAUUUUCGAGUUGUUUUUCACCAACUCGCCGACUUGGUGGUUGUGCUAGAAGUUGAUUAUGCCCGACGAU .............--------...((((....((...((((((((............((((((........))))))(((((((....))))).)).))))))))...)).....)))). ( -22.10) >DroEre_CAF1 182485 120 + 1 UCAUUGCAUUUUUUUUUUUUUUUUUUGUAUCAAUUUGCAAUUUUUUCGCUCAUUUUCAAGUUGUUUUUCACCAACUUGCCGACUUGGUGGUUGUGCAAAACGUUGAUUAUACCCGACGUU ...((((((...............(((((......))))).....((((.((..(((((((((........)))))))..))..))))))..))))))(((((((........))))))) ( -23.90) >DroYak_CAF1 147871 111 + 1 UCAUUGCAUUUUU---------UUUUGUAUCAAUUUGCAAUUUUUGUGCCCAUUUUCAAGUUGUUUUUCACCAACUUGCCGACUUGGUGGUUGUGCAAAACGUUGAUUAUACCCGAGGAU .....((((....---------..(((((......))))).....))))..((((((((((((........))))))(((((((....))))).))..................)))))) ( -20.80) >consensus UCAUUGCAUUUUU________UUUUCGUAUCAAUUUGCAAUUUUUUCGCUUAUUUUCGAGUUGUUUUUCACCAACUCGCCGACUUGGUGGUUGUGCUAGAAGUUGAUUAUGCCCGACGAU ........................((((.........((((((((............((((((........))))))(((((((....))))).)).))))))))..........)))). (-18.39 = -18.23 + -0.16)

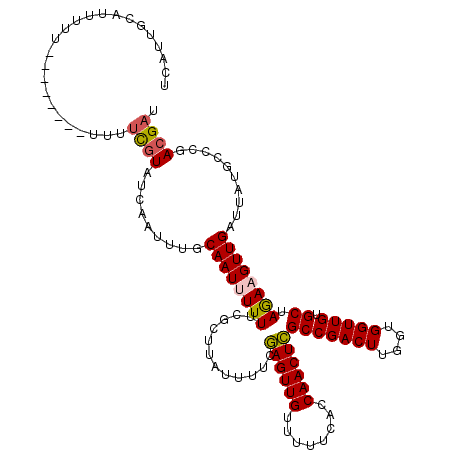
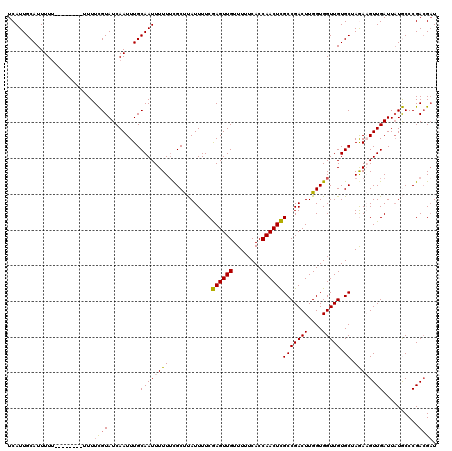
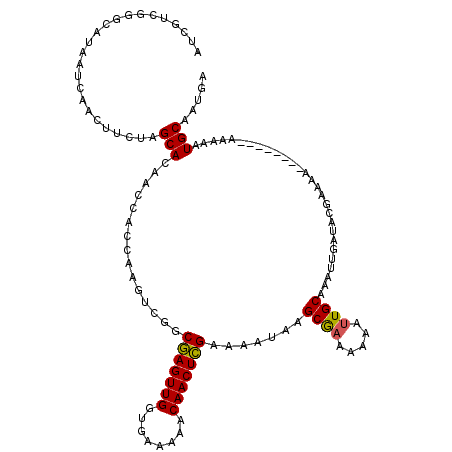
| Location | 20,708,099 – 20,708,209 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.80 |
| Mean single sequence MFE | -18.60 |
| Consensus MFE | -15.42 |
| Energy contribution | -14.98 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20708099 110 - 20766785 AUCGUCGGGCAUAAUCAACUUCUAGCACAACCACCAAGUCGGCGAGUUGGUGAAAAACAACUCGAAAAUAAGCGAAAAAAUUGCAAAUUGAUACGAAA----------AAAAUGCAAUGA .((((...((((.......(((..((.(.((......)).)))((((((........))))))))).....((((.....))))..............----------...)))).)))) ( -18.50) >DroSec_CAF1 141844 113 - 1 AUCGUCGGGCA-AAUCAACUUCUAGCACAACCACCAAGUCGGCGAGUUGGUGAAAAACAACUCGAAAAUAAGCGAAAAAAUUGCAAAUUGAUACGAAAAA------AAAAAAUGCAAUGA ...........-............(((...........(((.(((((((........))))))).......((((.....)))).........)))....------......)))..... ( -18.20) >DroSim_CAF1 146843 112 - 1 AUCGUCGGGCAUAAUCAACUUCUAGCACAACCACCAAGUCGGCGAGUUGGUGAAAAACAACUCGAAAAUAAGCGAAAAAAUUGCAAAUUGAUACGAAAA--------AAAAAUGCAAUGA .((((...((((.......(((..((.(.((......)).)))((((((........))))))))).....((((.....))))...............--------....)))).)))) ( -18.50) >DroEre_CAF1 182485 120 - 1 AACGUCGGGUAUAAUCAACGUUUUGCACAACCACCAAGUCGGCAAGUUGGUGAAAAACAACUUGAAAAUGAGCGAAAAAAUUGCAAAUUGAUACAAAAAAAAAAAAAAAAAAUGCAAUGA (((((.((......)).)))))(((((...........(((.(((((((........)))))))....)))((((.....))))............................)))))... ( -20.00) >DroYak_CAF1 147871 111 - 1 AUCCUCGGGUAUAAUCAACGUUUUGCACAACCACCAAGUCGGCAAGUUGGUGAAAAACAACUUGAAAAUGGGCACAAAAAUUGCAAAUUGAUACAAAA---------AAAAAUGCAAUGA (((....)))............(((((...(((((.....))(((((((........)))))))....)))(((.......)))..............---------.....)))))... ( -17.80) >consensus AUCGUCGGGCAUAAUCAACUUCUAGCACAACCACCAAGUCGGCGAGUUGGUGAAAAACAACUCGAAAAUAAGCGAAAAAAUUGCAAAUUGAUACGAAAA________AAAAAUGCAAUGA ........................(((...............(((((((........))))))).......((((.....))))............................)))..... (-15.42 = -14.98 + -0.44)

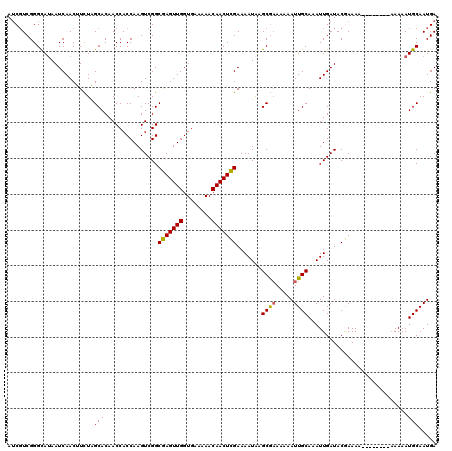
| Location | 20,708,129 – 20,708,249 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -20.59 |
| Consensus MFE | -18.12 |
| Energy contribution | -17.40 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20708129 120 - 20766785 AAAAUGAAAACUGCCAAACACAACCCUGCCCCUUCAUUAAAUCGUCGGGCAUAAUCAACUUCUAGCACAACCACCAAGUCGGCGAGUUGGUGAAAAACAACUCGAAAAUAAGCGAAAAAA ..........................(((((...(........)..))))).............((.......((.....))(((((((........))))))).......))....... ( -19.50) >DroSec_CAF1 141878 119 - 1 AAAAUGAAAACUGCCAAACACAACCCUGCCCCUUCAUUAAAUCGUCGGGCA-AAUCAACUUCUAGCACAACCACCAAGUCGGCGAGUUGGUGAAAAACAACUCGAAAAUAAGCGAAAAAA ..........................(((((...(........)..)))))-............((.......((.....))(((((((........))))))).......))....... ( -19.70) >DroSim_CAF1 146875 120 - 1 AAAAUGAAAACUGCCAAACACAACCCUGCCCCUUCAUUAAAUCGUCGGGCAUAAUCAACUUCUAGCACAACCACCAAGUCGGCGAGUUGGUGAAAAACAACUCGAAAAUAAGCGAAAAAA ..........................(((((...(........)..))))).............((.......((.....))(((((((........))))))).......))....... ( -19.50) >DroEre_CAF1 182525 114 - 1 -----GAAAACUGCAAAACACAACCCUGCCCCUU-AUUAAAACGUCGGGUAUAAUCAACGUUUUGCACAACCACCAAGUCGGCAAGUUGGUGAAAAACAACUUGAAAAUGAGCGAAAAAA -----......((((((((.......(((((...-...........)))))........))))))))...........(((.(((((((........)))))))....)))......... ( -22.20) >DroYak_CAF1 147902 120 - 1 AAAAUGAAAACUGCCAAACACAACCCUGCCCCUUCAUUAAAUCCUCGGGUAUAAUCAACGUUUUGCACAACCACCAAGUCGGCAAGUUGGUGAAAAACAACUUGAAAAUGGGCACAAAAA ...........((((...........(((((...............))))).......((((((.........((.....))(((((((........)))))))))))))))))...... ( -22.06) >consensus AAAAUGAAAACUGCCAAACACAACCCUGCCCCUUCAUUAAAUCGUCGGGCAUAAUCAACUUCUAGCACAACCACCAAGUCGGCGAGUUGGUGAAAAACAACUCGAAAAUAAGCGAAAAAA ..........................(((((...............))))).............((.......((.....))(((((((........))))))).......))....... (-18.12 = -17.40 + -0.72)

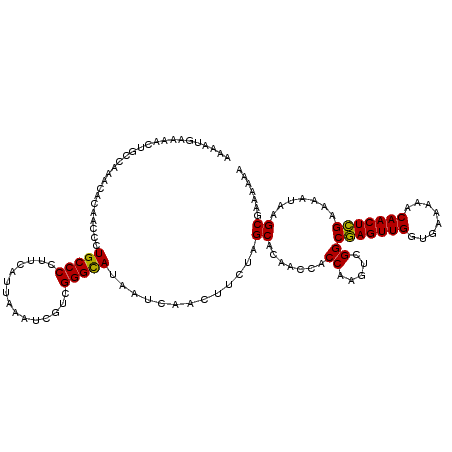
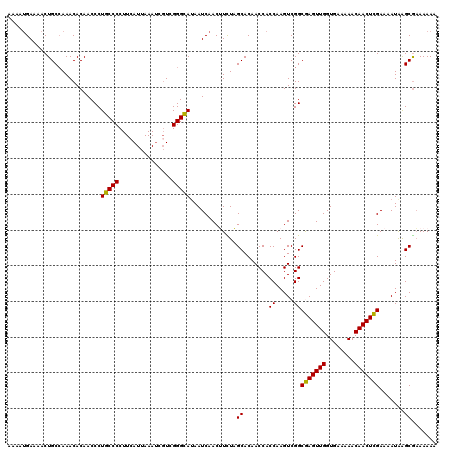
| Location | 20,708,209 – 20,708,317 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -25.47 |
| Energy contribution | -25.46 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20708209 108 + 20766785 UUAAUGAAGGGGCAGGGUUGUGUUUGGCAGUUUUCAUUUUAUGCGCUGCAU-GCCGU-----GCGUGACAUGAAAAACGGAUUUUUCGAUAU--UUUUAAUUCUGCUCUCUUUUGU .....(((((((((((((((.(((((....(((((((.((((((((.(...-.).))-----)))))).))))))).)))))..........--...))))))))).))))))... ( -32.44) >DroSec_CAF1 141957 116 + 1 UUAAUGAAGGGGCAGGGUUGUGUUUGGCAGUUUUCAUUUUAUGCGCUGCAUUGCCGUGCGGUGCGUGACAUGAAAAACGGAUUUUUGGAUUUUUUUUUAAUUUUGCUCUCUUUUGU .....(((((((((((((((.(((((....(((((((.(((((((((((((....))))))))))))).))))))).)))))...............))))))))).))))))... ( -38.29) >DroSim_CAF1 146955 111 + 1 UUAAUGAAGGGGCAGGGUUGUGUUUGGCAGUUUUCAUUUUAUGCGCUGCAUUGCCGU-----GCGUGACAUGAAAAACGGAUUUUUGGAUUUUUUUUUAAUUUUGCUCUCUUUUGU .....(((((((((((((((.(((((....(((((((.((((((((.((...)).))-----)))))).))))))).)))))...............))))))))).))))))... ( -30.69) >DroYak_CAF1 147982 109 + 1 UUAAUGAAGGGGCAGGGUUGUGUUUGGCAGUUUUCAUUUUAUGCGACGCAUUGCCGU-----GCGUGACAUGAAAAACGGAUUUUUAGAUUU--UUUUAAUUCUGCUCACUUAUGU ......((((((((((((((.((((((.(((((...(((((((..((((((....))-----))))..)))))))...))))).))))))..--...))))))))))).))).... ( -31.20) >consensus UUAAUGAAGGGGCAGGGUUGUGUUUGGCAGUUUUCAUUUUAUGCGCUGCAUUGCCGU_____GCGUGACAUGAAAAACGGAUUUUUGGAUUU__UUUUAAUUCUGCUCUCUUUUGU .....(((((((((((((((.(((((....(((((((.((((((...((......)).....)))))).))))))).)))))...............))))))))))).))))... (-25.47 = -25.46 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:19 2006