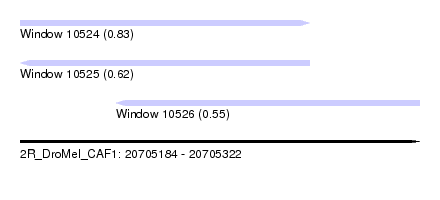
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,705,184 – 20,705,322 |
| Length | 138 |
| Max. P | 0.833086 |

| Location | 20,705,184 – 20,705,284 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.49 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -22.96 |
| Energy contribution | -23.40 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20705184 100 + 20766785 ACGGGGUGGC-------UUGAUGCCCAAAAGGGGUGGACAUUAUGCAUAUAUGUAUA------------UGUCUAACAGAGAGGGCAAUAUGCAUAAUGCUAUUCGUAAGGGUUGACCA- ...(((((..-------....))))).....((((((.((((((((((((.(((...------------(.((.....)).)..)))))))))))))))))))))....((.....)).- ( -32.50) >DroSec_CAF1 138813 97 + 1 ACGGGGUGGC-------UUGAUGCCGAAAAGGGGUGGACAUUAUGCA----UGUAUAC-----------UGUCUAAGAGGGAGGGCAAUAUGCAUAAAGCUAUCCGUAAGGGUUGACCA- ...(((((((-------((.((.((.....)).)).))..(((((((----(((...(-----------(.(((.....))).))..)))))))))).)))))))....((.....)).- ( -31.30) >DroSim_CAF1 142528 101 + 1 ACGGGGUGGC-------UUGAUGCCGAAAAGGGGUGGACAUUAUGCAUAUAUGUAUAC-----------UGUCUAAGAGGGAGGGCAAUAUGCAUAAAGCUAUCCGUAAGGGUUGACCA- ...(((((((-------((.((.((.....)).)).))..((((((((((.(((...(-----------(.((.....)).)).))))))))))))).)))))))....((.....)).- ( -32.90) >DroEre_CAF1 179542 117 + 1 ACGGGGUGGCUGAAUACUUGAUGCCGAAAAGGGGUGGACAUUAUGCAUAUACAUAUAUAUGA---UGGGUGUCUGAGAGGGAGGGUAAUAUGCAUAACGCUAUUCGUAAGGGUUGACCAG ...(((((((.......((.((.((.....)).)).))..((((((((((.(((....))).---......(((....)))......)))))))))).)))))))....((.....)).. ( -26.70) >DroYak_CAF1 144723 116 + 1 ACGGGGUGGCUGA---CUUGAUGCCGAAAAGGGGUGGACAUUAUGCAUAUAUGUAUACUGGGUCCUUGAUAUCUAAGAGGGAGGGCAAUAUGCAUAACGCUAUUCGUAAGGGUUGACCA- ...(((((((...---.((.((.((.....)).)).))..((((((((((.(((...((...(((((.........))))))).))))))))))))).)))))))....((.....)).- ( -30.30) >consensus ACGGGGUGGC_______UUGAUGCCGAAAAGGGGUGGACAUUAUGCAUAUAUGUAUAC___________UGUCUAAGAGGGAGGGCAAUAUGCAUAAAGCUAUUCGUAAGGGUUGACCA_ ...(((((((.......((.((.((.....)).)).))..((((((((((...................(((((........))))))))))))))).)))))))....((.....)).. (-22.96 = -23.40 + 0.44)

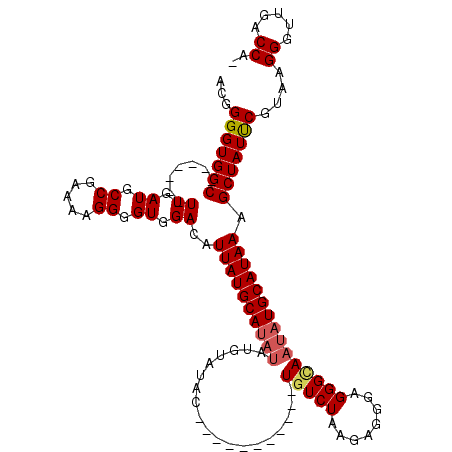
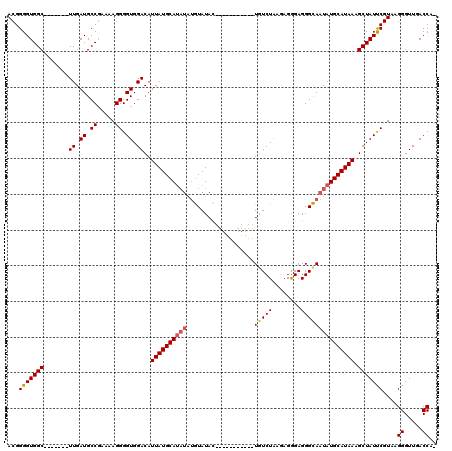
| Location | 20,705,184 – 20,705,284 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.49 |
| Mean single sequence MFE | -20.06 |
| Consensus MFE | -14.35 |
| Energy contribution | -15.27 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20705184 100 - 20766785 -UGGUCAACCCUUACGAAUAGCAUUAUGCAUAUUGCCCUCUCUGUUAGACA------------UAUACAUAUAUGCAUAAUGUCCACCCCUUUUGGGCAUCAA-------GCCACCCCGU -((((...............(((((((((((((((.....((.....))..------------....)).)))))))))))))....(((....)))......-------))))...... ( -20.80) >DroSec_CAF1 138813 97 - 1 -UGGUCAACCCUUACGGAUAGCUUUAUGCAUAUUGCCCUCCCUCUUAGACA-----------GUAUACA----UGCAUAAUGUCCACCCCUUUUCGGCAUCAA-------GCCACCCCGU -.(((..........(((((...((((((((..(((..((.......))..-----------)))...)----))))))))))))..........(((.....-------)))))).... ( -20.50) >DroSim_CAF1 142528 101 - 1 -UGGUCAACCCUUACGGAUAGCUUUAUGCAUAUUGCCCUCCCUCUUAGACA-----------GUAUACAUAUAUGCAUAAUGUCCACCCCUUUUCGGCAUCAA-------GCCACCCCGU -.(((..........(((((...(((((((((((((..((.......))..-----------))).....)))))))))))))))..........(((.....-------)))))).... ( -20.90) >DroEre_CAF1 179542 117 - 1 CUGGUCAACCCUUACGAAUAGCGUUAUGCAUAUUACCCUCCCUCUCAGACACCCA---UCAUAUAUAUGUAUAUGCAUAAUGUCCACCCCUUUUCGGCAUCAAGUAUUCAGCCACCCCGU ..(((...............(((((((((((((......................---............)))))))))))))............(((............)))))).... ( -17.68) >DroYak_CAF1 144723 116 - 1 -UGGUCAACCCUUACGAAUAGCGUUAUGCAUAUUGCCCUCCCUCUUAGAUAUCAAGGACCCAGUAUACAUAUAUGCAUAAUGUCCACCCCUUUUCGGCAUCAAG---UCAGCCACCCCGU -.(((...............(((((((((((((((..((...((((.(....).))))...))....)).)))))))))))))............(((......---...)))))).... ( -20.40) >consensus _UGGUCAACCCUUACGAAUAGCGUUAUGCAUAUUGCCCUCCCUCUUAGACA___________GUAUACAUAUAUGCAUAAUGUCCACCCCUUUUCGGCAUCAA_______GCCACCCCGU ..(((...............(((((((((((((.....................................)))))))))))))............(((............)))))).... (-14.35 = -15.27 + 0.92)

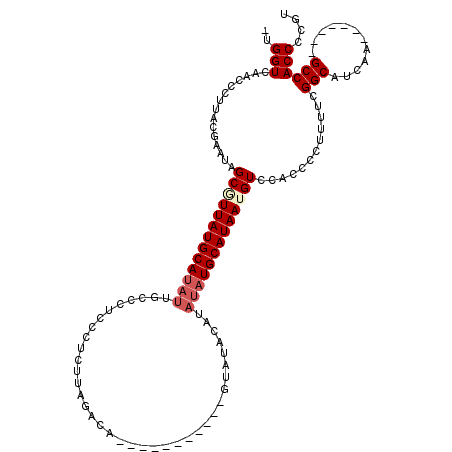
| Location | 20,705,217 – 20,705,322 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.53 |
| Mean single sequence MFE | -20.56 |
| Consensus MFE | -6.15 |
| Energy contribution | -6.75 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.30 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
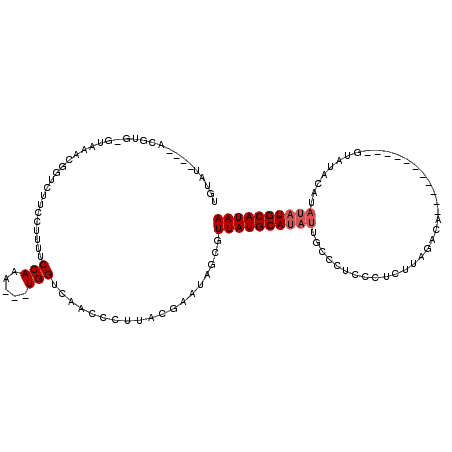
>2R_DroMel_CAF1 20705217 105 - 20766785 UGUAUAUGUACGUGUGUCAACGGUCUUCCAUUUCCAAA---UGGUCAACCCUUACGAAUAGCAUUAUGCAUAUUGCCCUCUCUGUUAGACA------------UAUACAUAUAUGCAUAA (((((((((..(((((((((((((...(((((....))---)))...)))......((((((.....)).)))).........))).))))------------)))..)))))))))... ( -26.30) >DroSec_CAF1 138846 98 - 1 UGUAU----ACGUGCGUAAACGGUCUUCCUUUUCCAAG---UGGUCAACCCUUACGGAUAGCUUUAUGCAUAUUGCCCUCCCUCUUAGACA-----------GUAUACA----UGCAUAA (((((----((((((((((..(((..(((......(((---.((....)))))..)))..)))))))))))......((.......))...-----------)))))))----....... ( -20.00) >DroSim_CAF1 142561 102 - 1 UGUAU----ACGUGUGUAAACGGUCUUUCUUUUCCAAG---UGGUCAACCCUUACGGAUAGCUUUAUGCAUAUUGCCCUCCCUCUUAGACA-----------GUAUACAUAUAUGCAUAA (((((----(..((((((....((((.........(((---.((....)))))..(((..((..((....))..))..))).....)))).-----------.))))))..))))))... ( -23.50) >DroEre_CAF1 179582 102 - 1 UGUAU----AUG-----------UCGUCCUUUUCCAAACCCUGGUCAACCCUUACGAAUAGCGUUAUGCAUAUUACCCUCCCUCUCAGACACCCA---UCAUAUAUAUGUAUAUGCAUAA .(((.----..(-----------(.(.((.............)).).))...))).....((((.((((((((......................---......)))))))))))).... ( -12.51) >DroYak_CAF1 144760 109 - 1 UAUUU----AUGU----AUGCUCUCGUCCUUUUCCAAA---UGGUCAACCCUUACGAAUAGCGUUAUGCAUAUUGCCCUCCCUCUUAGAUAUCAAGGACCCAGUAUACAUAUAUGCAUAA (((.(----((((----(((((...((((((.((.((.---.((.........(((.....)))...((.....))....))..)).))....))))))..)))))))))).)))..... ( -20.50) >consensus UGUAU____ACGUG_GUAAACGGUCUUCCUUUUCCAAA___UGGUCAACCCUUACGAAUAGCGUUAUGCAUAUUGCCCUCCCUCUUAGACA___________GUAUACAUAUAUGCAUAA .................................(((.....)))...................((((((((((.....................................)))))))))) ( -6.15 = -6.75 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:07 2006