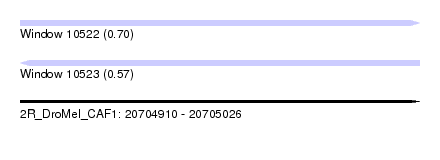
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,704,910 – 20,705,026 |
| Length | 116 |
| Max. P | 0.702187 |

| Location | 20,704,910 – 20,705,026 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.37 |
| Mean single sequence MFE | -37.14 |
| Consensus MFE | -23.70 |
| Energy contribution | -24.54 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.702187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20704910 116 + 20766785 AAAGACGAUUUGAGUAGGAUGCUUCUGGCACGCGACGAUUUGUAUUUAAGAAAUAGGUUUUGGGAGUGUGCCAAGAAUUUAGCGUGCCAUCAGCCUCUCAGCUUGAUCUUAUUUGU---- .(((((((.(((((.(((.((....((((((((.....((((....))))...((((((((.((......)).)))))))))))))))).)).)))))))).))).))))......---- ( -33.60) >DroSec_CAF1 138550 116 + 1 AAAGAGGAUUCGAGUAGGAUGCUUCUGGCACGCGACGAUUUGUAUUUAAGAAAUAGGUUUUGGGAGUGUGCCAAGUAUUUAGCGUGGCAUCAGCCUCUCUGCUCGAUCUUAUUUGU---- ....(((((.(((((((((((((..((((((((..(((..(.((((.....)))).)..)))...))))))))))))))....(.(((....))).).))))))))))))......---- ( -40.70) >DroSim_CAF1 142265 116 + 1 AAAGAGGAUUCGAGUAGGAUGCUUCUGGCACGCGACGAUUUUUAUUUAAGAAAUAGGUUUUGGGAGUGUGCCAAGUAUUUAGCGUGGCAUCAGCCUCUCUGCUCGAUCUUAUUUGU---- ....(((((.(((((((((((((..((((((((..(((..((((((.....))))))..)))...))))))))))))))....(.(((....))).).))))))))))))......---- ( -41.40) >DroEre_CAF1 179297 116 + 1 AAAGACAAUUCGAGGAGGAUGCUCCUGGCACGCGACGAGUUGAGUUUAAGAAUUGGGAUUUGGGAGUGUGCCAAGAGUUUAGCGUGCCAUCAGCCUCUGCCC----AUUUUUUUGUUUUU ((((((((...((((.(((.(((..(((((((((((.......)))((((..((((.((........)).))))...))))))))))))..))).)))..))----.))...)))))))) ( -35.20) >DroYak_CAF1 144482 113 + 1 AAAG---AUUCGAUAAGGAUGCUCCAGGCACGCGACGAGUUGAGUUUAAGAAUUAGGAUUUGGGAGUGUGCCAAGAUUUUAGCGUGCCAUCAGCCUCUGCGCUCGAAUUUUUUUGU---- ((((---((((((...(((.(((...((((((((((.......))).......((((((((.((......)).)))))))))))))))...))).)))....))))))))))....---- ( -34.80) >consensus AAAGACGAUUCGAGUAGGAUGCUUCUGGCACGCGACGAUUUGUAUUUAAGAAAUAGGUUUUGGGAGUGUGCCAAGAAUUUAGCGUGCCAUCAGCCUCUCCGCUCGAUCUUAUUUGU____ .........((((((.(((.(((..((((((((..(.((((.........)))).)..(((((.......)))))......))))))))..))).)))..)))))).............. (-23.70 = -24.54 + 0.84)

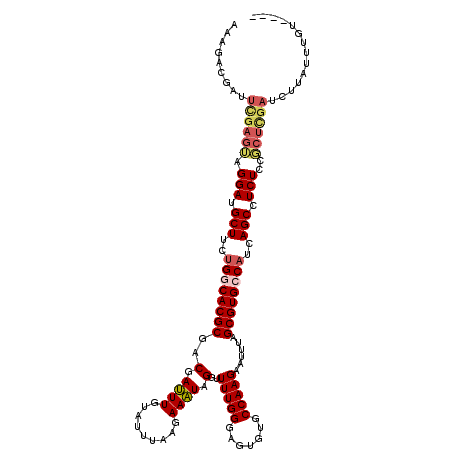
| Location | 20,704,910 – 20,705,026 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.37 |
| Mean single sequence MFE | -25.69 |
| Consensus MFE | -17.84 |
| Energy contribution | -20.04 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20704910 116 - 20766785 ----ACAAAUAAGAUCAAGCUGAGAGGCUGAUGGCACGCUAAAUUCUUGGCACACUCCCAAAACCUAUUUCUUAAAUACAAAUCGUCGCGUGCCAGAAGCAUCCUACUCAAAUCGUCUUU ----......(((((.....(((((((.((.((((((((.......((((.......))))....((((.....)))).........))))))))....)).))).))))....))))). ( -27.50) >DroSec_CAF1 138550 116 - 1 ----ACAAAUAAGAUCGAGCAGAGAGGCUGAUGCCACGCUAAAUACUUGGCACACUCCCAAAACCUAUUUCUUAAAUACAAAUCGUCGCGUGCCAGAAGCAUCCUACUCGAAUCCUCUUU ----........(((((((.((.((.(((..((.(((((.......((((.......))))....((((.....)))).........))))).))..))).)))).)))).)))...... ( -25.10) >DroSim_CAF1 142265 116 - 1 ----ACAAAUAAGAUCGAGCAGAGAGGCUGAUGCCACGCUAAAUACUUGGCACACUCCCAAAACCUAUUUCUUAAAUAAAAAUCGUCGCGUGCCAGAAGCAUCCUACUCGAAUCCUCUUU ----........(((((((.((.((.(((..((.(((((.......((((.......))))....((((.....)))).........))))).))..))).)))).)))).)))...... ( -25.40) >DroEre_CAF1 179297 116 - 1 AAAAACAAAAAAAU----GGGCAGAGGCUGAUGGCACGCUAAACUCUUGGCACACUCCCAAAUCCCAAUUCUUAAACUCAACUCGUCGCGUGCCAGGAGCAUCCUCCUCGAAUUGUCUUU ..............----(((((((((..(((((....)....((((((((((.(................................).))))))))))))))..))))....))))).. ( -26.25) >DroYak_CAF1 144482 113 - 1 ----ACAAAAAAAUUCGAGCGCAGAGGCUGAUGGCACGCUAAAAUCUUGGCACACUCCCAAAUCCUAAUUCUUAAACUCAACUCGUCGCGUGCCUGGAGCAUCCUUAUCGAAU---CUUU ----........((((((.....((.(((...(((((((.......((((.......))))..............((.......)).)))))))...))).))....))))))---.... ( -24.20) >consensus ____ACAAAUAAGAUCGAGCAGAGAGGCUGAUGGCACGCUAAAUUCUUGGCACACUCCCAAAACCUAUUUCUUAAAUACAAAUCGUCGCGUGCCAGAAGCAUCCUACUCGAAUCCUCUUU ..............(((((.((.((.(((..((((((((.......((((.......))))..............((.......)).))))))))..))).)))).)))))......... (-17.84 = -20.04 + 2.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:05 2006