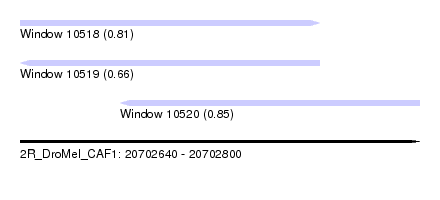
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,702,640 – 20,702,800 |
| Length | 160 |
| Max. P | 0.848243 |

| Location | 20,702,640 – 20,702,760 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -27.16 |
| Energy contribution | -27.04 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.24 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
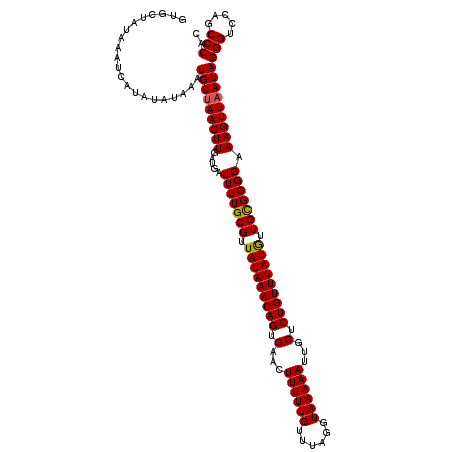
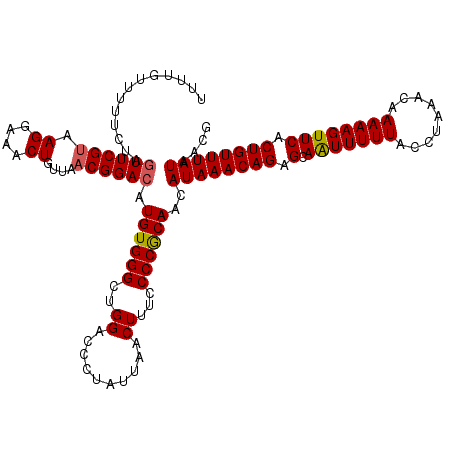
>2R_DroMel_CAF1 20702640 120 + 20766785 GUGCUAUAAAUCAUAUAUAAAUGUAAACUUGAUGAUUUUGCGUUAUAAACAGUGAACUUUUUGUUUAGGUAAAAAUUGCUCUGUUUAUGUUGCGGGGAAAGUUAAUAGGGUCCAGCCCAC .....................(((.(((((.....(((((((.((((((((((((((.....)))))((((.....))))))))))))).))))))).))))).)))(((.....))).. ( -24.40) >DroSec_CAF1 136241 120 + 1 GUGCUAUAAAUCAUAUAUAAAUGUUAACUUGAUGAUUUUGCGUUAUAAACAGUGAACUUUUUGUUUAGGUAAAAAUUGCUCUGUUUAUGUUGCGGGGAAAGUUAAUAGGGUCCAGCCCAC .....................(((((((((.....(((((((.((((((((((((((.....)))))((((.....))))))))))))).))))))).)))))))))(((.....))).. ( -28.30) >DroSim_CAF1 139876 120 + 1 GUGCUAUAAAUCAUAUAUAAAUGUUAACUUGAUGAUUUUGCGUUAUAAACAGUGAACUUUUUGUUUAGGUAAAAAUUGCUCUGUUUAUGUUGCGGGGAAAGUUAAUAGGGUCCAGCCCAC .....................(((((((((.....(((((((.((((((((((((((.....)))))((((.....))))))))))))).))))))).)))))))))(((.....))).. ( -28.30) >DroEre_CAF1 176936 120 + 1 GUGCUAUAAAUCAUAUAUAAAUGUUAACUUGAUGAUUUUGCGUUAUAAACAGCGAACUUUUUGUUUAGGUAAAAACUGCUCUGUUUAUGUUGUGGGGAAAGUUAAUAGGGUCCAGCCCAC .....................(((((((((.....(((..((.(((((((((.(...((((((......))))))...).))))))))).))..))).)))))))))(((.....))).. ( -26.30) >DroYak_CAF1 141691 120 + 1 GUGCUAUAAAUCAUAUAUAAAUGUUAACUUGAUGAUUUUGCGUUAUAAACAGUGAACUUUUUGUUUAUGUAAAAACUGCUCUGUUUAUAUUGCGGGGAAAGUUAAUAGGGUCCAGCCCAC .....................(((((((((.....(((((((.(((((((((.(...((((((......))))))...).))))))))).))))))).)))))))))(((.....))).. ( -28.30) >consensus GUGCUAUAAAUCAUAUAUAAAUGUUAACUUGAUGAUUUUGCGUUAUAAACAGUGAACUUUUUGUUUAGGUAAAAAUUGCUCUGUUUAUGUUGCGGGGAAAGUUAAUAGGGUCCAGCCCAC .....................(((((((((.....(((((((.(((((((((.(...((((((......))))))...).))))))))).))))))).)))))))))(((.....))).. (-27.16 = -27.04 + -0.12)

| Location | 20,702,640 – 20,702,760 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -19.21 |
| Consensus MFE | -18.39 |
| Energy contribution | -17.99 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20702640 120 - 20766785 GUGGGCUGGACCCUAUUAACUUUCCCCGCAACAUAAACAGAGCAAUUUUUACCUAAACAAAAAGUUCACUGUUUAUAACGCAAAAUCAUCAAGUUUACAUUUAUAUAUGAUUUAUAGCAC (((((..((..........))...)))))...((((((((.(.(((((((.........)))))))).))))))))...((.(((((((.................)))))))...)).. ( -19.03) >DroSec_CAF1 136241 120 - 1 GUGGGCUGGACCCUAUUAACUUUCCCCGCAACAUAAACAGAGCAAUUUUUACCUAAACAAAAAGUUCACUGUUUAUAACGCAAAAUCAUCAAGUUAACAUUUAUAUAUGAUUUAUAGCAC (((((..((..........))...)))))...((((((((.(.(((((((.........)))))))).))))))))...((.(((((((.................)))))))...)).. ( -19.03) >DroSim_CAF1 139876 120 - 1 GUGGGCUGGACCCUAUUAACUUUCCCCGCAACAUAAACAGAGCAAUUUUUACCUAAACAAAAAGUUCACUGUUUAUAACGCAAAAUCAUCAAGUUAACAUUUAUAUAUGAUUUAUAGCAC (((((..((..........))...)))))...((((((((.(.(((((((.........)))))))).))))))))...((.(((((((.................)))))))...)).. ( -19.03) >DroEre_CAF1 176936 120 - 1 GUGGGCUGGACCCUAUUAACUUUCCCCACAACAUAAACAGAGCAGUUUUUACCUAAACAAAAAGUUCGCUGUUUAUAACGCAAAAUCAUCAAGUUAACAUUUAUAUAUGAUUUAUAGCAC (((((..((..........))...)))))...((((((((.(.(.(((((........))))).).).))))))))...((.(((((((.................)))))))...)).. ( -19.23) >DroYak_CAF1 141691 120 - 1 GUGGGCUGGACCCUAUUAACUUUCCCCGCAAUAUAAACAGAGCAGUUUUUACAUAAACAAAAAGUUCACUGUUUAUAACGCAAAAUCAUCAAGUUAACAUUUAUAUAUGAUUUAUAGCAC (((((..((..........))...)))))..(((((((((.(.(.(((((........))))).).).)))))))))..((.(((((((.................)))))))...)).. ( -19.73) >consensus GUGGGCUGGACCCUAUUAACUUUCCCCGCAACAUAAACAGAGCAAUUUUUACCUAAACAAAAAGUUCACUGUUUAUAACGCAAAAUCAUCAAGUUAACAUUUAUAUAUGAUUUAUAGCAC (((((..((..........))...)))))...((((((((.(.(((((((.........)))))))).))))))))...((.(((((((.................)))))))...)).. (-18.39 = -17.99 + -0.40)


| Location | 20,702,680 – 20,702,800 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -23.92 |
| Energy contribution | -23.92 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
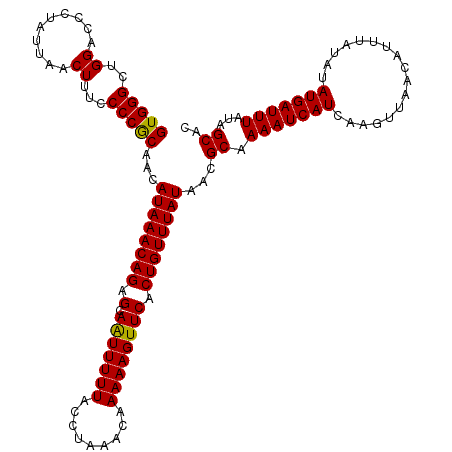
>2R_DroMel_CAF1 20702680 120 - 20766785 UUUUGUUUUUCUAGUUCGUAAGGAAACUGUUAACGGACAUGUGGGCUGGACCCUAUUAACUUUCCCCGCAACAUAAACAGAGCAAUUUUUACCUAAACAAAAAGUUCACUGUUUAUAACG .............((((((.((....))....)))))).((((((..((..........))...))))))..((((((((.(.(((((((.........)))))))).)))))))).... ( -26.40) >DroSec_CAF1 136281 120 - 1 UUUUGUUUUUCUUGUUCGUAAGGAAACUGUUAACGGACAUGUGGGCUGGACCCUAUUAACUUUCCCCGCAACAUAAACAGAGCAAUUUUUACCUAAACAAAAAGUUCACUGUUUAUAACG ............(((((((.((....))....)))))))((((((..((..........))...))))))..((((((((.(.(((((((.........)))))))).)))))))).... ( -26.90) >DroSim_CAF1 139916 120 - 1 UUUUGUUUUUCUAUUUCGUAAGGAAACUGUUAACGGACAUGUGGGCUGGACCCUAUUAACUUUCCCCGCAACAUAAACAGAGCAAUUUUUACCUAAACAAAAAGUUCACUGUUUAUAACG ..............(((((.((....))....)))))..((((((..((..........))...))))))..((((((((.(.(((((((.........)))))))).)))))))).... ( -22.50) >DroEre_CAF1 176976 120 - 1 UUUUGUUUUUCUAGUUCGAAAGGAAACUGUUAACGGACAUGUGGGCUGGACCCUAUUAACUUUCCCCACAACAUAAACAGAGCAGUUUUUACCUAAACAAAAAGUUCGCUGUUUAUAACG .............(((((..((....)).....))))).((((((..((..........))...))))))..((((((((.(.(.(((((........))))).).).)))))))).... ( -24.70) >DroYak_CAF1 141731 119 - 1 -UUUGUUUUUCUAGUUCGUAAGGAAACUGUUAACGGACAUGUGGGCUGGACCCUAUUAACUUUCCCCGCAAUAUAAACAGAGCAGUUUUUACAUAAACAAAAAGUUCACUGUUUAUAACG -............((((((.((....))....)))))).((((((..((..........))...)))))).(((((((((.(.(.(((((........))))).).).)))))))))... ( -27.10) >consensus UUUUGUUUUUCUAGUUCGUAAGGAAACUGUUAACGGACAUGUGGGCUGGACCCUAUUAACUUUCCCCGCAACAUAAACAGAGCAAUUUUUACCUAAACAAAAAGUUCACUGUUUAUAACG .............((((((.((....))....)))))).((((((..((..........))...))))))..((((((((.(.(((((((.........)))))))).)))))))).... (-23.92 = -23.92 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:02 2006