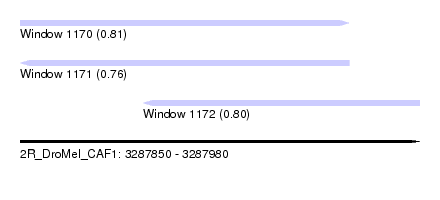
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,287,850 – 3,287,980 |
| Length | 130 |
| Max. P | 0.813190 |

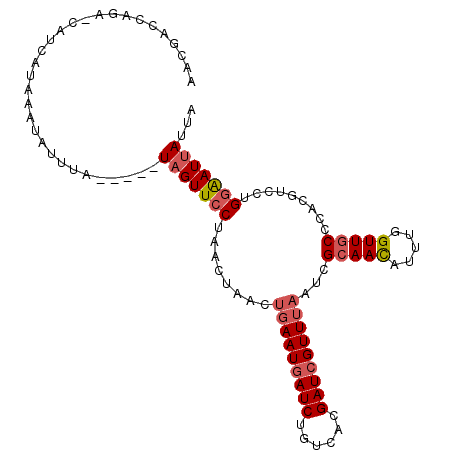
| Location | 3,287,850 – 3,287,957 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -19.15 |
| Consensus MFE | -13.96 |
| Energy contribution | -15.02 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
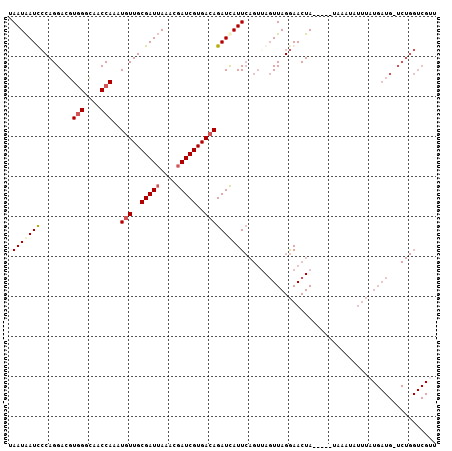
>2R_DroMel_CAF1 3287850 107 + 20766785 ACCGACCAGAUCAUCAUAAAUAUUUAUACAAUAGUUCCUAACUAACUGAAUGAUCUGUCACGAUCGUUUAAUCGCAACAUUUGGUUGCCCACGUCCUGGGAUUAUUA ..(((((((((....................((((.....))))..(((((((((......)))))))))........)))))))))((((.....))))....... ( -23.10) >DroSec_CAF1 56278 102 + 1 AACGACCAGAACAUCAUAAAUAUUUA-----UAGUUCCUAACUAACUGAAUAAUCUGUCACGAUCGUUUAAUCGCAACAUUUGGUUGCCCACGUCCUGGAAUUAUUA ..((((((((..........((((((-----((((.....))))..))))))...(((..((((......))))..))))))))))).(((.....)))........ ( -17.00) >DroSim_CAF1 66817 102 + 1 AACGACCAGAGCAUCAUAAAUAUUUA-----UAGUUCCUAACUCACUGAAUGAUCUGUCACGAUCGUUUAAUCGCAACAUUUGGUUGCCCACGUCCUGGAAUUAUUA .....((((..(..............-----...............(((((((((......)))))))))...(((((.....)))))....)..))))........ ( -22.30) >DroYak_CAF1 55188 81 + 1 A---------------U--UUAUUUA-----UAGUUUCUAAGUA----AAUGAUCUGUCACGAUCGUUUUAUCGCAAUAUUUGGUUUCCUAAUUCCUGCGAUCAUUA (---------------(--(((((((-----.......))))))----)))((((......)))).....((((((....((((....))))....))))))..... ( -14.20) >consensus AACGACCAGA_CAUCAUAAAUAUUUA_____UAGUUCCUAACUAACUGAAUGAUCUGUCACGAUCGUUUAAUCGCAACAUUUGGUUGCCCACGUCCUGGAAUUAUUA ...............................(((((((........(((((((((......)))))))))...(((((.....))))).........)))))))... (-13.96 = -15.02 + 1.06)

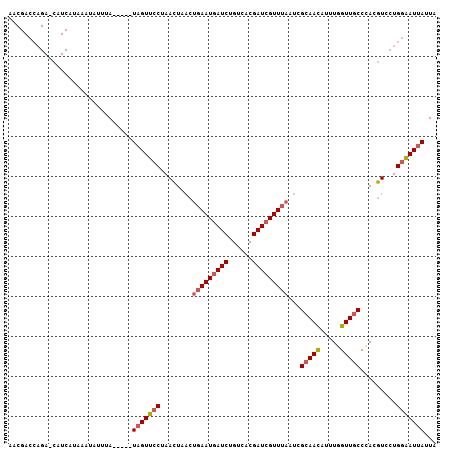
| Location | 3,287,850 – 3,287,957 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -23.22 |
| Consensus MFE | -14.80 |
| Energy contribution | -15.68 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
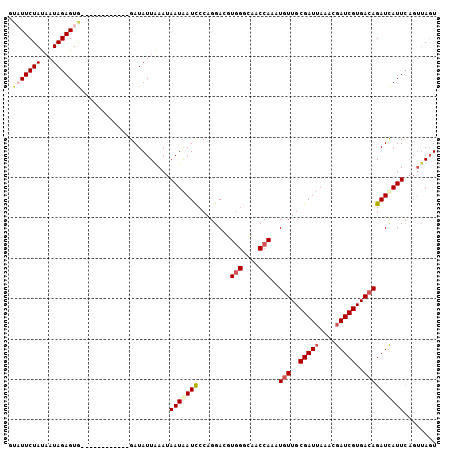
>2R_DroMel_CAF1 3287850 107 - 20766785 UAAUAAUCCCAGGACGUGGGCAACCAAAUGUUGCGAUUAAACGAUCGUGACAGAUCAUUCAGUUAGUUAGGAACUAUUGUAUAAAUAUUUAUGAUGAUCUGGUCGGU ...(((((.(((.(..(((....)))..).))).)))))....((((...(((((((((((((.(((.....))))))).............)))))))))..)))) ( -26.51) >DroSec_CAF1 56278 102 - 1 UAAUAAUUCCAGGACGUGGGCAACCAAAUGUUGCGAUUAAACGAUCGUGACAGAUUAUUCAGUUAGUUAGGAACUA-----UAAAUAUUUAUGAUGUUCUGGUCGUU ........(((((((((((....))...(((..(((((....)))))..)))(((((......)))))........-----............)))))))))..... ( -27.40) >DroSim_CAF1 66817 102 - 1 UAAUAAUUCCAGGACGUGGGCAACCAAAUGUUGCGAUUAAACGAUCGUGACAGAUCAUUCAGUGAGUUAGGAACUA-----UAAAUAUUUAUGAUGCUCUGGUCGUU ........((((..(.(((....)))..(((..(((((....)))))..))).(((((.........(((...)))-----.........))))))..))))..... ( -24.17) >DroYak_CAF1 55188 81 - 1 UAAUGAUCGCAGGAAUUAGGAAACCAAAUAUUGCGAUAAAACGAUCGUGACAGAUCAUU----UACUUAGAAACUA-----UAAAUAA--A---------------U .....(((((((......(....)......))))))).....((((......))))...----.............-----.......--.---------------. ( -14.80) >consensus UAAUAAUCCCAGGACGUGGGCAACCAAAUGUUGCGAUUAAACGAUCGUGACAGAUCAUUCAGUUAGUUAGGAACUA_____UAAAUAUUUAUGAUG_UCUGGUCGUU .(((((((........(((....)))..(((..(((((....)))))..))))))))))................................................ (-14.80 = -15.68 + 0.87)

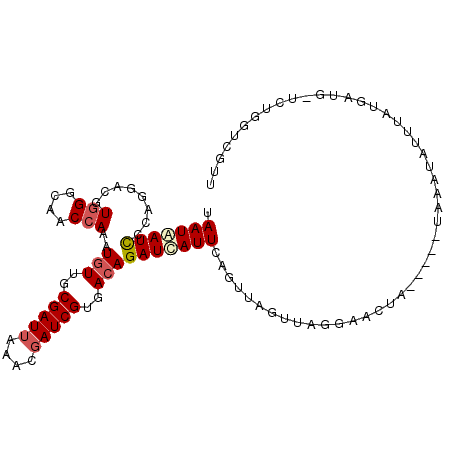
| Location | 3,287,890 – 3,287,980 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 81.67 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -15.45 |
| Energy contribution | -16.95 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
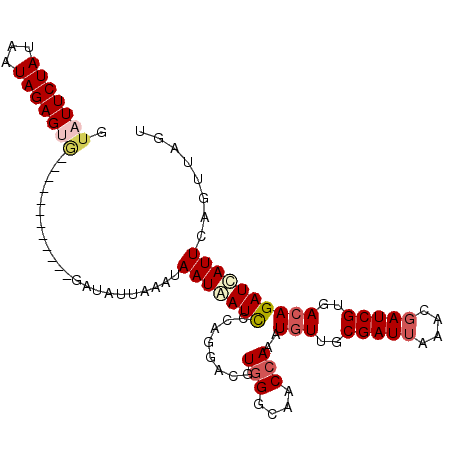
>2R_DroMel_CAF1 3287890 90 - 20766785 ---UUCUAU-AUAGAGUG------------GAUAUUAAAUAAUAAUCCCAGGACGUGGGCAACCAAAUGUUGCGAUUAAACGAUCGUGACAGAUCAUUCAGUUAGU ---......-...(((((------------(........................(((....)))..(((..(((((....)))))..)))..))))))....... ( -21.80) >DroSec_CAF1 56313 94 - 1 GUAUUCUAUAAUAGAGUG------------GAUAUUAAAUAAUAAUUCCAGGACGUGGGCAACCAAAUGUUGCGAUUAAACGAUCGUGACAGAUUAUUCAGUUAGU .(((((((...)))))))------------...(((((..(((((((........(((....)))..(((..(((((....)))))..))))))))))...))))) ( -22.20) >DroSim_CAF1 66852 94 - 1 GUAUUCUAUAAUAGAGUG------------GAUAUUAAAUAAUAAUUCCAGGACGUGGGCAACCAAAUGUUGCGAUUAAACGAUCGUGACAGAUCAUUCAGUGAGU .............(((((------------(........................(((....)))..(((..(((((....)))))..)))..))))))....... ( -21.80) >DroYak_CAF1 55206 102 - 1 AUUUUCUACAAUAGAGUAAAAUUGUCCAGCGAUAUUGAAUAAUGAUCGCAGGAAUUAGGAAACCAAAUAUUGCGAUAAAACGAUCGUGACAGAUCAUU----UACU ..............((((((.(((((..(((((...........(((((((......(....)......)))))))......))))))))))....))----)))) ( -19.73) >consensus GUAUUCUAUAAUAGAGUG____________GAUAUUAAAUAAUAAUCCCAGGACGUGGGCAACCAAAUGUUGCGAUUAAACGAUCGUGACAGAUCAUUCAGUUAGU .(((((((...)))))))......................(((((((........(((....)))..(((..(((((....)))))..))))))))))........ (-15.45 = -16.95 + 1.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:01 2006