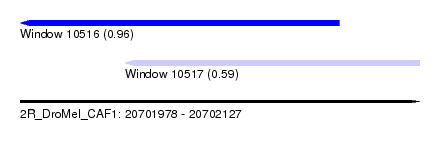
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,701,978 – 20,702,127 |
| Length | 149 |
| Max. P | 0.963814 |

| Location | 20,701,978 – 20,702,097 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.15 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -24.70 |
| Energy contribution | -25.58 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20701978 119 - 20766785 UCGCCGGCAGAUGUAGAGCAUUUAAGUACAGAAAGUACGCAGAUAUGCAUAAUCCUUAUACACACGUUUUACUUAGACAAUGGUAAGAUUUGAUUAGGGAUGCCGGGUGUUGAAAACAC- ...((((((((((.....)))))..((((.....))))(((....)))...(((((((....((.((((((((........)))))))).))..))))))))))))((((.....))))- ( -33.80) >DroSec_CAF1 135506 118 - 1 U--CCGGCAGAUGUACAGCAUGCAAGUACAGAAAGUACACCGAUAUGCAUAAUCCUUAUACACGCAUUUUACUUAGACAAUGGUAAGAUUUGAUUAAGGAUGCCGGGUGUUGAAAAUAUA (--(((((.........((((((..((((.....))))...).)))))...(((((((....((.((((((((........)))))))).))..)))))))))))))............. ( -31.60) >DroSim_CAF1 139177 116 - 1 U--CCGGCAGAUGUACAGCAUGCAAGUACAGAAAGUACGCCGAUAUGCAUAAUCCUUA--CACACAUUUUACUUAGACAAUGGUAAGAUUUGAUUAAGGAUGCCGGGUGUUGAAAAUAUA (--(((((.........((((((..((((.....))))...).)))))...(((((((--..((.((((((((........)))))))).))..)))))))))))))............. ( -34.00) >DroEre_CAF1 176186 120 - 1 GCCACGGCAGAUGUACAACAGUUAAGUACAGAAAGUACGCCGAUAUGCAUAAUCCUUAUGCACACAUUUUACUUAGACAAUAGUAAGAUUUGAUUAAGGAUGACGGGUGUUGAAAAAAUA (((.((((...((.....)).....((((.....)))))))).........(((((((....((.((((((((........)))))))).))..)))))))....)))............ ( -27.40) >DroYak_CAF1 140302 120 - 1 GCGCAGGCAGAUGUACAGCAGCUAGGUACAGAAAGUACGCCGAUAUGCAUAAUCCUUAUGCACACAUUUUACUUAGACAAUGGUAAGAUUUGAUUAAGGAUGACGGGUGUUGAAAAAAUA (((...((...(((((.........)))))....)).)))((((((.(...(((((((....((.((((((((........)))))))).))..)))))))...).))))))........ ( -27.80) >consensus UC_CCGGCAGAUGUACAGCAUGUAAGUACAGAAAGUACGCCGAUAUGCAUAAUCCUUAUACACACAUUUUACUUAGACAAUGGUAAGAUUUGAUUAAGGAUGCCGGGUGUUGAAAAAAUA .....((((((((.....)))))..((((.....)))))))(((((.(...(((((((....((.((((((((........)))))))).))..)))))))...).)))))......... (-24.70 = -25.58 + 0.88)

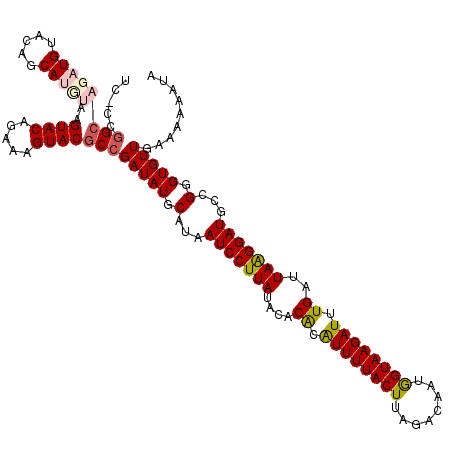
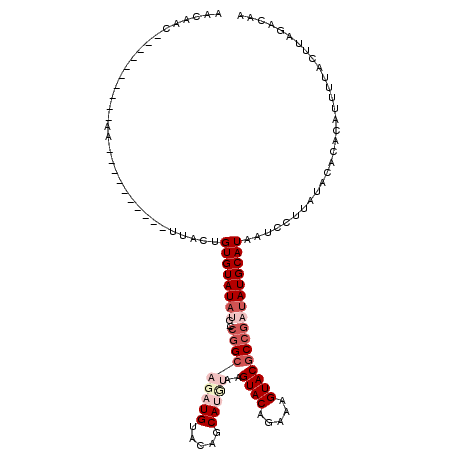
| Location | 20,702,017 – 20,702,127 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.00 |
| Mean single sequence MFE | -21.02 |
| Consensus MFE | -11.64 |
| Energy contribution | -13.96 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20702017 110 - 20766785 AACAACGUAGAAAUACAA----------UUAUGGUGUAUAUCGCCGGCAGAUGUAGAGCAUUUAAGUACAGAAAGUACGCAGAUAUGCAUAAUCCUUAUACACACGUUUUACUUAGACAA ......(((....)))..----------.....(((((((((....(((((((.....)))))..((((.....)))))).)))))))))...............((((.....)))).. ( -23.20) >DroSec_CAF1 135546 95 - 1 AACAAC----------AA-------------CUGUGUAUAU--CCGGCAGAUGUACAGCAUGCAAGUACAGAAAGUACACCGAUAUGCAUAAUCCUUAUACACGCAUUUUACUUAGACAA ......----------..-------------.(((((((((--(.((....((((.....)))).((((.....)))).))))))))))))............................. ( -16.70) >DroSim_CAF1 139217 93 - 1 AACAAC----------AA-------------CUGUGUAUAU--CCGGCAGAUGUACAGCAUGCAAGUACAGAAAGUACGCCGAUAUGCAUAAUCCUUA--CACACAUUUUACUUAGACAA ......----------..-------------.(((((((((--(.(((...((((.....)))).((((.....))))))))))))))))).......--.................... ( -21.20) >DroEre_CAF1 176226 99 - 1 ---------------------AAUUAUCUUACUGUGUAUUGCCACGGCAGAUGUACAACAGUUAAGUACAGAAAGUACGCCGAUAUGCAUAAUCCUUAUGCACACAUUUUACUUAGACAA ---------------------...((((..((((((((((((....)))...)))).)))))...((((.....))))...))))((((((.....)))))).................. ( -20.70) >DroYak_CAF1 140342 119 - 1 -ACAACGUAUAAAGACAAAUAAAUUAUCUUACGGUGUAUAGCGCAGGCAGAUGUACAGCAGCUAGGUACAGAAAGUACGCCGAUAUGCAUAAUCCUUAUGCACACAUUUUACUUAGACAA -.....((...(((.................((((((((.((....))...(((((.........)))))....))))))))...((((((.....)))))).........)))..)).. ( -23.30) >consensus AACAAC__________AA__________UUACUGUGUAUAUC_CCGGCAGAUGUACAGCAUGUAAGUACAGAAAGUACGCCGAUAUGCAUAAUCCUUAUACACACAUUUUACUUAGACAA .................................((((((((...(((((((((.....)))))..((((.....)))))))))))))))).............................. (-11.64 = -13.96 + 2.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:59 2006