
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,700,431 – 20,700,589 |
| Length | 158 |
| Max. P | 0.998539 |

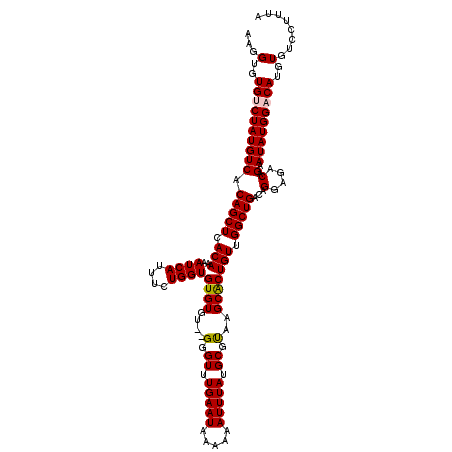
| Location | 20,700,431 – 20,700,549 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -27.66 |
| Energy contribution | -27.74 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20700431 118 + 20766785 AAGGUGUGUCUAUGUCACAGCUCACAAAAUCAUUUCUGGUGUGUGU--GGGUUUGAAUAAAAAAUUUAUGCGUAAGCACUGUUGGCUGACAGGAGACAGAAUAUGGACAUGUGUCCCUUA (((((((((((((((((.((((((((..((((....))))...)))--))))))))............(((....)))(((((..((....)).)))))..))))))))))....)))). ( -34.00) >DroSec_CAF1 134327 117 + 1 -AGGUGUGUCUAUGUCACAGCUCACAAAAUCAUUUCUGGUGUGUGU--GGGUUUGAAUAAAAAAUUUAUGCGUAAGCACUGUUGGCUGACAGGAGACAGAAUAUGGACAUGUGUCCUUUA -(((.((((((((((((.((((((((..((((....))))...)))--))))))))............(((....)))(((((..((....)).)))))..)))))))))....)))... ( -33.60) >DroSim_CAF1 136869 118 + 1 AAGGUGUGUCUAUGUCACAGCUCACAAAAUCAUUUCUGGUGUGUGU--GGGUUUGAAUAAAAAAUUUAUGCGUAAGCACUGUUGGCUGACAGGAGACAGAAUAUGGACAUGUGUCCUUUA ((((.((((((((((((.((((((((..((((....))))...)))--))))))))............(((....)))(((((..((....)).)))))..)))))))))....)))).. ( -34.50) >DroEre_CAF1 174841 113 + 1 ---GUGUGCCUAUGUCACAGCUCACAAAAUCAUUUCUGGUGUGU----GGGUUUGAAUAAAAAAUUUAUGCGUUAGCACUGUUGGCUGACAGGAGACAGAAUAUGGACAUGUGUCCUUUA ---.(((.(((...(((.((((((((..((((....)))).)))----))))))))...............((((((.(....))))))))))..)))......((((....)))).... ( -29.40) >DroYak_CAF1 139191 120 + 1 AAGGUGUGCCUAUGUCACAGCUCACAAAAUCAUUUCUGGUGUGUGUGGGGGUUUGAAUAAAAAAUUUAUGCGCUGGCGCUGUUGGCUGACAGGAGACAGAAUAUGGACAUGUGUCCUUUA ((((...((.((((((.(((((.(((..((((....))))..((((.((.((.(((((.....))))).)).)).))))))).)))))...(....)........)))))).)))))).. ( -32.90) >consensus AAGGUGUGUCUAUGUCACAGCUCACAAAAUCAUUUCUGGUGUGUGU__GGGUUUGAAUAAAAAAUUUAUGCGUAAGCACUGUUGGCUGACAGGAGACAGAAUAUGGACAUGUGUCCUUUA ...(..((((((((((.(((((.(((..((((....))))((((....(.((.(((((.....))))).)).)..))))))).)))))...(....).).)))))))))..)........ (-27.66 = -27.74 + 0.08)

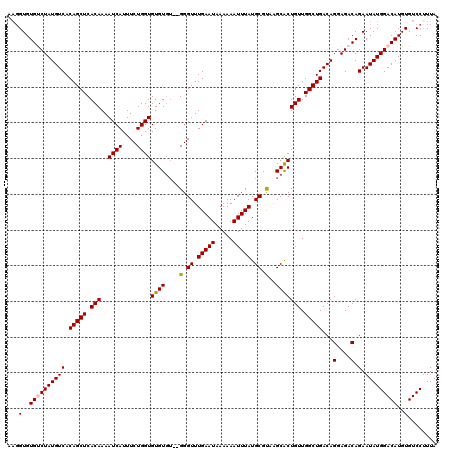
| Location | 20,700,431 – 20,700,549 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -24.31 |
| Consensus MFE | -23.64 |
| Energy contribution | -23.24 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20700431 118 - 20766785 UAAGGGACACAUGUCCAUAUUCUGUCUCCUGUCAGCCAACAGUGCUUACGCAUAAAUUUUUUAUUCAAACCC--ACACACACCAGAAAUGAUUUUGUGAGCUGUGACAUAGACACACCUU ....((((....))))......(((((..(((((((.....((((....))))...................--.((((...((....))....)))).))..))))).)))))...... ( -23.70) >DroSec_CAF1 134327 117 - 1 UAAAGGACACAUGUCCAUAUUCUGUCUCCUGUCAGCCAACAGUGCUUACGCAUAAAUUUUUUAUUCAAACCC--ACACACACCAGAAAUGAUUUUGUGAGCUGUGACAUAGACACACCU- ....((((....))))......(((((..(((((((.....((((....))))...................--.((((...((....))....)))).))..))))).))))).....- ( -23.90) >DroSim_CAF1 136869 118 - 1 UAAAGGACACAUGUCCAUAUUCUGUCUCCUGUCAGCCAACAGUGCUUACGCAUAAAUUUUUUAUUCAAACCC--ACACACACCAGAAAUGAUUUUGUGAGCUGUGACAUAGACACACCUU ....((((....))))......(((((..(((((((.....((((....))))...................--.((((...((....))....)))).))..))))).)))))...... ( -23.90) >DroEre_CAF1 174841 113 - 1 UAAAGGACACAUGUCCAUAUUCUGUCUCCUGUCAGCCAACAGUGCUAACGCAUAAAUUUUUUAUUCAAACCC----ACACACCAGAAAUGAUUUUGUGAGCUGUGACAUAGGCACAC--- ....((((....))))......(((((..(((((((.....((((....))))..................(----(((...((....))....)))).))..))))).)))))...--- ( -25.10) >DroYak_CAF1 139191 120 - 1 UAAAGGACACAUGUCCAUAUUCUGUCUCCUGUCAGCCAACAGCGCCAGCGCAUAAAUUUUUUAUUCAAACCCCCACACACACCAGAAAUGAUUUUGUGAGCUGUGACAUAGGCACACCUU ....((((....))))......(((((..(((((((.....))..((((.((((((.((....(((..................)))..)).)))))).))))))))).)))))...... ( -24.97) >consensus UAAAGGACACAUGUCCAUAUUCUGUCUCCUGUCAGCCAACAGUGCUUACGCAUAAAUUUUUUAUUCAAACCC__ACACACACCAGAAAUGAUUUUGUGAGCUGUGACAUAGACACACCUU ....((((....))))......(((((..((((.....(((((((....))..........................((((.((....))....)))).))))))))).)))))...... (-23.64 = -23.24 + -0.40)


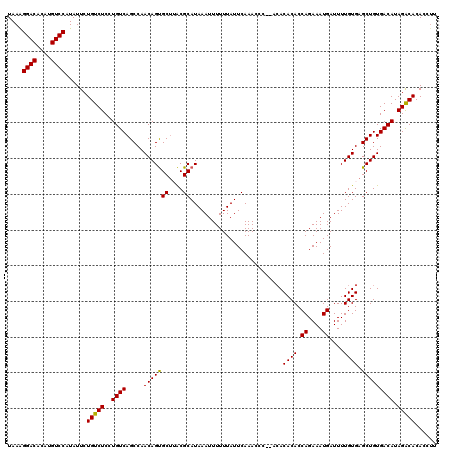
| Location | 20,700,471 – 20,700,589 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.29 |
| Mean single sequence MFE | -27.42 |
| Consensus MFE | -24.76 |
| Energy contribution | -24.52 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20700471 118 + 20766785 GUGUGU--GGGUUUGAAUAAAAAAUUUAUGCGUAAGCACUGUUGGCUGACAGGAGACAGAAUAUGGACAUGUGUCCCUUAUAACAACUGACAGAAGCGUAUAGCUUGAGUAUAAAAAGUU ..((((--((((((.......))))))))))(((..(.((((..(.((...(....).......((((....)))).......)).)..))))((((.....)))))..)))........ ( -26.50) >DroSec_CAF1 134366 118 + 1 GUGUGU--GGGUUUGAAUAAAAAAUUUAUGCGUAAGCACUGUUGGCUGACAGGAGACAGAAUAUGGACAUGUGUCCUUUAUAACAACUGACAGAAGCGUAUAGCUUGAGUAUAAAAAGUU ..((((--((((((.......))))))))))(((..(.((((..(.((...(....).......((((....)))).......)).)..))))((((.....)))))..)))........ ( -26.90) >DroSim_CAF1 136909 118 + 1 GUGUGU--GGGUUUGAAUAAAAAAUUUAUGCGUAAGCACUGUUGGCUGACAGGAGACAGAAUAUGGACAUGUGUCCUUUAUAACAACUGACAGAAGCGUAUAGCUUGAGUAUAAAAAGUU ..((((--((((((.......))))))))))(((..(.((((..(.((...(....).......((((....)))).......)).)..))))((((.....)))))..)))........ ( -26.90) >DroEre_CAF1 174878 116 + 1 GUGU----GGGUUUGAAUAAAAAAUUUAUGCGUUAGCACUGUUGGCUGACAGGAGACAGAAUAUGGACAUGUGUCCUUUAUAACAACUGACAGAAGCGCAUAGUUUGGGUAUAAAAAGUU ....----((.(((..(((...((.(((((((((....((((..(.((...(....).......((((....)))).......)).)..)))).))))))))).))...)))..))).)) ( -26.00) >DroYak_CAF1 139231 120 + 1 GUGUGUGGGGGUUUGAAUAAAAAAUUUAUGCGCUGGCGCUGUUGGCUGACAGGAGACAGAAUAUGGACAUGUGUCCUUUAUAACAACUGACAGAAGCGCAUAGUUUGGGUAUAAAAAGUU ........((.(((..(((...((.(((((((((....((((..(.((...(....).......((((....)))).......)).)..)))).))))))))).))...)))..))).)) ( -30.80) >consensus GUGUGU__GGGUUUGAAUAAAAAAUUUAUGCGUAAGCACUGUUGGCUGACAGGAGACAGAAUAUGGACAUGUGUCCUUUAUAACAACUGACAGAAGCGUAUAGCUUGAGUAUAAAAAGUU ........................(((((((.......((((..(.((...(....).......((((....)))).......)).)..))))((((.....))))..)))))))..... (-24.76 = -24.52 + -0.24)


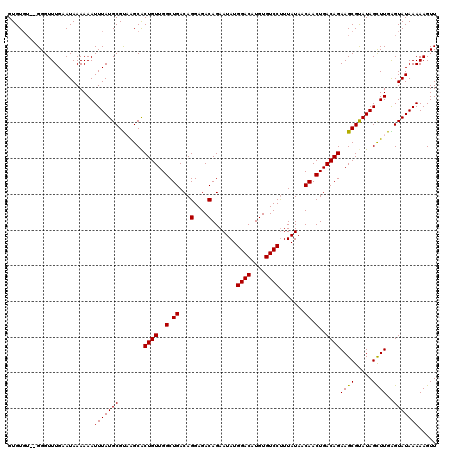
| Location | 20,700,471 – 20,700,589 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.29 |
| Mean single sequence MFE | -19.02 |
| Consensus MFE | -15.08 |
| Energy contribution | -14.92 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20700471 118 - 20766785 AACUUUUUAUACUCAAGCUAUACGCUUCUGUCAGUUGUUAUAAGGGACACAUGUCCAUAUUCUGUCUCCUGUCAGCCAACAGUGCUUACGCAUAAAUUUUUUAUUCAAACCC--ACACAC ..........((..((((.....))))..))..((((.......((((....)))).....(((.(....).))).)))).((((....))))...................--...... ( -17.20) >DroSec_CAF1 134366 118 - 1 AACUUUUUAUACUCAAGCUAUACGCUUCUGUCAGUUGUUAUAAAGGACACAUGUCCAUAUUCUGUCUCCUGUCAGCCAACAGUGCUUACGCAUAAAUUUUUUAUUCAAACCC--ACACAC ..........((..((((.....))))..))..((((.......((((....)))).....(((.(....).))).)))).((((....))))...................--...... ( -17.40) >DroSim_CAF1 136909 118 - 1 AACUUUUUAUACUCAAGCUAUACGCUUCUGUCAGUUGUUAUAAAGGACACAUGUCCAUAUUCUGUCUCCUGUCAGCCAACAGUGCUUACGCAUAAAUUUUUUAUUCAAACCC--ACACAC ..........((..((((.....))))..))..((((.......((((....)))).....(((.(....).))).)))).((((....))))...................--...... ( -17.40) >DroEre_CAF1 174878 116 - 1 AACUUUUUAUACCCAAACUAUGCGCUUCUGUCAGUUGUUAUAAAGGACACAUGUCCAUAUUCUGUCUCCUGUCAGCCAACAGUGCUAACGCAUAAAUUUUUUAUUCAAACCC----ACAC ..................((((((...((((..((((......((((.(((.(........)))).))))..))))..))))......))))))..................----.... ( -19.00) >DroYak_CAF1 139231 120 - 1 AACUUUUUAUACCCAAACUAUGCGCUUCUGUCAGUUGUUAUAAAGGACACAUGUCCAUAUUCUGUCUCCUGUCAGCCAACAGCGCCAGCGCAUAAAUUUUUUAUUCAAACCCCCACACAC ..................((((((((.......((((((.....((((....)))).....(((.(....).)))..))))))...)))))))).......................... ( -24.10) >consensus AACUUUUUAUACUCAAGCUAUACGCUUCUGUCAGUUGUUAUAAAGGACACAUGUCCAUAUUCUGUCUCCUGUCAGCCAACAGUGCUUACGCAUAAAUUUUUUAUUCAAACCC__ACACAC .......................((..((((..((((......((((.(((.(........)))).))))..))))..)))).))................................... (-15.08 = -14.92 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:57 2006