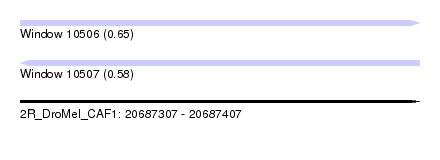
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,687,307 – 20,687,407 |
| Length | 100 |
| Max. P | 0.646060 |

| Location | 20,687,307 – 20,687,407 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
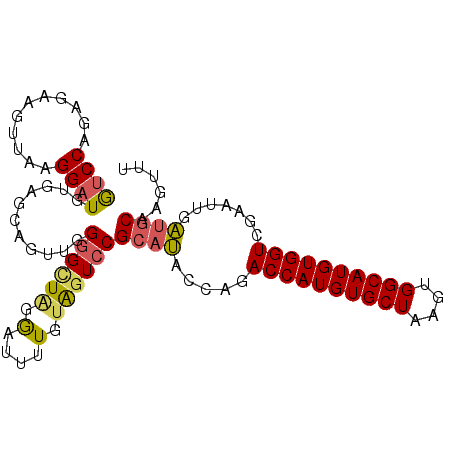
| Reading direction | forward |
| Mean pairwise identity | 83.00 |
| Mean single sequence MFE | -18.55 |
| Consensus MFE | -14.89 |
| Energy contribution | -14.08 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20687307 100 + 20766785 AAACUUGCAUCAAUUUGACCACAUGCCACUUAGCACAUGGUCUGGUAUGCGGACUACAAAACCCUAGCCCCAACUGCUGACAUCCUUAACUUCUCUGGAC ....((((((..((..(((((..(((......)))..)))))..))))))))............((((.......))))...(((...........))). ( -18.30) >DroSec_CAF1 120808 100 + 1 AAACUUGCAUCAAUUCGACCACAUGCCACUUAGCACAUGGUCUGGUAUGCGGACUACAAAAUCCUAGCCCCAACUGCUCACAUCCUUAACUUCUCUGGAC ......(((.......(((((..(((......)))..)))))(((...(((((........)))..)).)))..))).....(((...........))). ( -17.90) >DroSim_CAF1 122284 100 + 1 AAACUUGCAUCAAUUCGACCACAUGCCACUUAGCACAUGGUCUGGUAUGCGGACUACAAAAUCCUAGCCCCAACUGCUCACAUCCUUAACUUCUCUGGAC ......(((.......(((((..(((......)))..)))))(((...(((((........)))..)).)))..))).....(((...........))). ( -17.90) >DroEre_CAF1 161505 99 + 1 AAACUUGCAUCAAUUCGACCACAUGCCACUUAGCACAUGGU-UGGUAUGCGGACUACAAAAUCCCAUCCCCAACCACUCACAUCCCCAACUUCUUUGGAC ......((((....(((((((..(((......)))..))))-))).))))(((........))).....................((((.....)))).. ( -18.50) >DroYak_CAF1 125297 100 + 1 AAACUUGCAUCAAUUCGACCACAUGCCACUUAGCACAUGGUUUGGUAUGCGGACUACAAAAUCCCAACCCCAACCACUCACAUCCCAAACUUCUUUGGAC ......((((..((..(((((..(((......)))..)))))..))))))(((........)))....................(((((....))))).. ( -17.10) >DroAna_CAF1 128424 89 + 1 AAACUUGUGGCCGCUUGACCACAUGCCGCUUAGCACAUGGUUUCGCCUUCGGACCCGACUACUCCAACGCCCA--UCUCGCAUCCU---------UGGGU ......((((..((..(((((..(((......)))..)))))..))..(((....)))))))......(((((--...........---------))))) ( -21.60) >consensus AAACUUGCAUCAAUUCGACCACAUGCCACUUAGCACAUGGUCUGGUAUGCGGACUACAAAAUCCCAGCCCCAACUGCUCACAUCCUUAACUUCUCUGGAC ....((((((..((..(((((..(((......)))..)))))..))))))))..............................(((...........))). (-14.89 = -14.08 + -0.80)

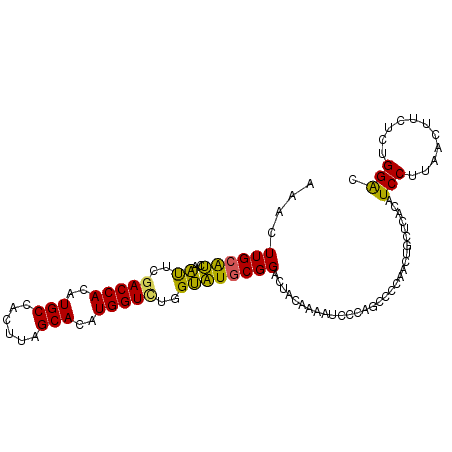
| Location | 20,687,307 – 20,687,407 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 83.00 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -21.09 |
| Energy contribution | -21.07 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20687307 100 - 20766785 GUCCAGAGAAGUUAAGGAUGUCAGCAGUUGGGGCUAGGGUUUUGUAGUCCGCAUACCAGACCAUGUGCUAAGUGGCAUGUGGUCAAAUUGAUGCAAGUUU ((((...........))))...(((.....((((((.(....).))))))((((....(((((((((((....)))))))))))......))))..))). ( -30.20) >DroSec_CAF1 120808 100 - 1 GUCCAGAGAAGUUAAGGAUGUGAGCAGUUGGGGCUAGGAUUUUGUAGUCCGCAUACCAGACCAUGUGCUAAGUGGCAUGUGGUCGAAUUGAUGCAAGUUU ((((...........))))..((((.....((((((.(....).))))))((((....(((((((((((....)))))))))))......))))..)))) ( -32.70) >DroSim_CAF1 122284 100 - 1 GUCCAGAGAAGUUAAGGAUGUGAGCAGUUGGGGCUAGGAUUUUGUAGUCCGCAUACCAGACCAUGUGCUAAGUGGCAUGUGGUCGAAUUGAUGCAAGUUU ((((...........))))..((((.....((((((.(....).))))))((((....(((((((((((....)))))))))))......))))..)))) ( -32.70) >DroEre_CAF1 161505 99 - 1 GUCCAAAGAAGUUGGGGAUGUGAGUGGUUGGGGAUGGGAUUUUGUAGUCCGCAUACCA-ACCAUGUGCUAAGUGGCAUGUGGUCGAAUUGAUGCAAGUUU .(((((.....)))))(((....((((((((..((((((((....))))).))).)))-)))))(((((....)))))...)))................ ( -29.00) >DroYak_CAF1 125297 100 - 1 GUCCAAAGAAGUUUGGGAUGUGAGUGGUUGGGGUUGGGAUUUUGUAGUCCGCAUACCAAACCAUGUGCUAAGUGGCAUGUGGUCGAAUUGAUGCAAGUUU .((((((....)))))).(((...(((((.(((((((((((....))))).)).)))..((((((((((....))))))))))).)))))..)))..... ( -28.00) >DroAna_CAF1 128424 89 - 1 ACCCA---------AGGAUGCGAGA--UGGGCGUUGGAGUAGUCGGGUCCGAAGGCGAAACCAUGUGCUAAGCGGCAUGUGGUCAAGCGGCCACAAGUUU .....---------.....((..(.--(((.((((.((((..(((....)))..))....(((((((((....))))))))))).)))).))))..)).. ( -27.90) >consensus GUCCAGAGAAGUUAAGGAUGUGAGCAGUUGGGGCUAGGAUUUUGUAGUCCGCAUACCAGACCAUGUGCUAAGUGGCAUGUGGUCGAAUUGAUGCAAGUUU ((((...........))))...........((((((.(....).))))))((((.....((((((((((....)))))))))).......))))...... (-21.09 = -21.07 + -0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:49 2006