
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,684,135 – 20,684,236 |
| Length | 101 |
| Max. P | 0.882347 |

| Location | 20,684,135 – 20,684,236 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 80.97 |
| Mean single sequence MFE | -33.85 |
| Consensus MFE | -23.51 |
| Energy contribution | -23.60 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20684135 101 - 20766785 GUCUUACUGACCU-AUUGCAGAUACUCGUCCUUUCUCUGAUUCUCUUCUGCGGCAGCUCACAUGCUGUUGUCAGUGAAUUGGCCCGUCAGAGCGAGUCUGCC (((.....)))..-...(((((..((((..((..(.(..((((....((((((((((......))))))).))).))))..)...)..))..))))))))). ( -34.90) >DroPse_CAF1 119859 100 - 1 GUA-AACUGGUUA-CUUGCAGAUACUCAUCGUGCUGCUGGCCCUGCUCCGUGGGAGCUCCAAUGCCAUUAUUAGCGAGCUAGCCCGCCAGGGCGAAAGUGCA ...-.....((.(-(((((((.(((.....)))))))..((((((....(((((((((((((((....)))).).)))))..)))))))))))..)))))). ( -33.90) >DroSec_CAF1 117516 101 - 1 GUCUUACUGGCCU-AUUGCAGAUACUCGUCCUUUCUCUGAUUCUCUUCUGUGGCAGCUCACAUGCUGUUAUCAGUGAGUUGGCCCGUCAGAGCGAGUCUGUC (((.....)))..-...(((((..((((..((..(.(..((((....((((((((((......))))))).))).))))..)...)..))..))))))))). ( -30.80) >DroSim_CAF1 118947 101 - 1 GUCUUACUGACCU-AUUGCAGAUACUCGUCCUUUCUCUGAUUCUCUUCUGCGGCAGCUCACAUGCUGUUGUCAGUGAGUUGGCCCGUCAGAGCGAGUCUGCC (((.....)))..-...(((((..((((..((..(.(..((((....((((((((((......))))))).))).))))..)...)..))..))))))))). ( -34.90) >DroEre_CAF1 158029 102 - 1 GUCUAACUGACCCGUUUGCAGAUUCUCGUCCUAUUUCUGAGCCUCUUCUGCGGCAGCUCACAUGCUGUUGUCAGUGAGUUGGCUCGUCAGGCCGAGUCUGUC (((.....)))......(((((..((((.(((.....((((((.(((((((((((((......))))))).))).)))..))))))..))).))))))))). ( -39.10) >DroYak_CAF1 122405 102 - 1 GUCUUAUUGAACCGUUUGCAGAUUCUCGUCCUUUCUCUGAUCCUCUUUAGCGGCAGCUCACAUGCUAUUGUCAGUGAGUUGGCCCGUCAGACCGAGUCUGCC .................((((((((..(((......((((......)))).((((((((((..((....))..))))))).))).....))).)))))))). ( -29.50) >consensus GUCUUACUGACCU_AUUGCAGAUACUCGUCCUUUCUCUGAUCCUCUUCUGCGGCAGCUCACAUGCUGUUGUCAGUGAGUUGGCCCGUCAGAGCGAGUCUGCC .................(((((..((((......(((((((((..(((.((((((((......))))))))....)))..))...)))))))))))))))). (-23.51 = -23.60 + 0.09)

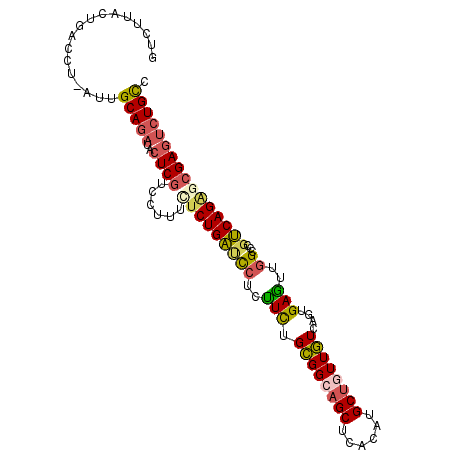
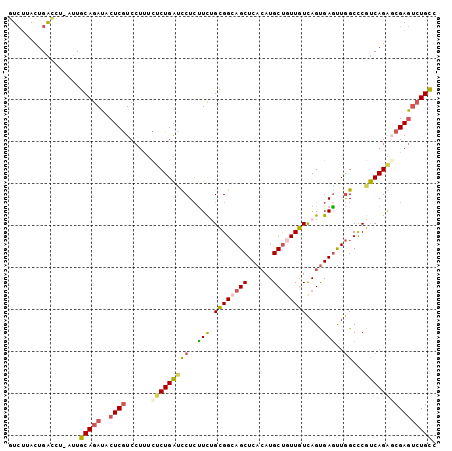
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:47 2006