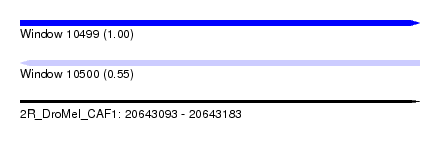
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,643,093 – 20,643,183 |
| Length | 90 |
| Max. P | 0.996848 |

| Location | 20,643,093 – 20,643,183 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 73.08 |
| Mean single sequence MFE | -29.86 |
| Consensus MFE | -14.22 |
| Energy contribution | -13.74 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
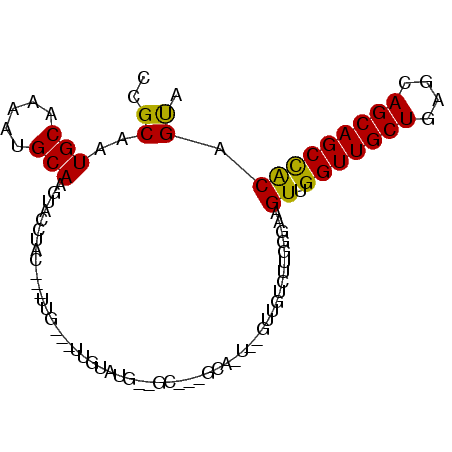
>2R_DroMel_CAF1 20643093 90 + 20766785 CCGCAAUGCAAAACGCAAGUACCUAC---UUG---UUGUAUGGAGCUGCGCACUUGGUUGUCUUGGGAAGUUGGUUGCUCAGCAGCAGCCACAGUA ((.(((.((((..((((.((.(((((---...---..))).)).)))))).......)))).)))))..((.(((((((....))))))))).... ( -26.20) >DroSec_CAF1 94254 73 + 1 CCGCAAUGCAAAAUGCAAGUACCUAC---UUG---UUGUAUG-----------------GUCUUGGGGAGUUGGUUGCUUAGCAGCAGCCACAGUA ((.(((.((.....((((((....))---)))---)......-----------------)).))).)).((.(((((((....))))))))).... ( -23.50) >DroSim_CAF1 95155 73 + 1 CCGCAAUGCAAAAUGCAAGUACCUAC---UUG---UUGUAUG-----------------GUCUUGGGGAGUUGGUUGCUUAGCAGCAGCCACAGUA ((.(((.((.....((((((....))---)))---)......-----------------)).))).)).((.(((((((....))))))))).... ( -23.50) >DroEre_CAF1 133640 88 + 1 CCGCAGUGCAAAAUGCAAGCACCUACAGCUUGCGCUUGUAUAGUGCACUGCACUGUGUUGU--------GUUGGUUGCUGAGCAGCAGCUACAGUA ..((((((((..((((((((.(.........).))))))))..))))))))(((((.....--------...(((((((....)))))))))))). ( -37.60) >DroYak_CAF1 98164 96 + 1 CCGCAGUGCAAAAUGCAGGCAUCUACAACGUGCACUUGUAUGAUGCUAUGCAUUUUGUUGUCUUGGGAAGUUGGUUGCUGAUCAGCAGCCGCAGCA (((.((.(((((((((((((((((((((.(....)))))).)))))).))))))))))...)))))...((.(((((((....))))))))).... ( -38.50) >consensus CCGCAAUGCAAAAUGCAAGUACCUAC___UUG___UUGUAUG__GC___GCA_U__GUUGUCUUGGGAAGUUGGUUGCUGAGCAGCAGCCACAGUA ..((..(((.....)))....................................................((.(((((((....))))))))).)). (-14.22 = -13.74 + -0.48)

| Location | 20,643,093 – 20,643,183 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 73.08 |
| Mean single sequence MFE | -23.96 |
| Consensus MFE | -10.52 |
| Energy contribution | -10.40 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
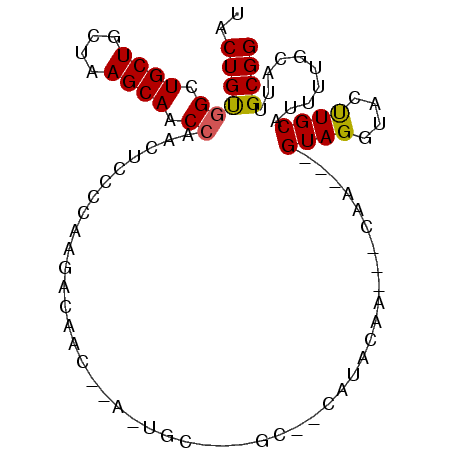
>2R_DroMel_CAF1 20643093 90 - 20766785 UACUGUGGCUGCUGCUGAGCAACCAACUUCCCAAGACAACCAAGUGCGCAGCUCCAUACAA---CAA---GUAGGUACUUGCGUUUUGCAUUGCGG ..((((((.((((....)))).)).......((((((...(((((((.(.(((........---..)---)).)))))))).))))))....)))) ( -21.50) >DroSec_CAF1 94254 73 - 1 UACUGUGGCUGCUGCUAAGCAACCAACUCCCCAAGAC-----------------CAUACAA---CAA---GUAGGUACUUGCAUUUUGCAUUGCGG .....(((.((((....)))).))).....(((((((-----------------(.(((..---...---)))))).)))(((........))))) ( -17.20) >DroSim_CAF1 95155 73 - 1 UACUGUGGCUGCUGCUAAGCAACCAACUCCCCAAGAC-----------------CAUACAA---CAA---GUAGGUACUUGCAUUUUGCAUUGCGG .....(((.((((....)))).))).....(((((((-----------------(.(((..---...---)))))).)))(((........))))) ( -17.20) >DroEre_CAF1 133640 88 - 1 UACUGUAGCUGCUGCUCAGCAACCAAC--------ACAACACAGUGCAGUGCACUAUACAAGCGCAAGCUGUAGGUGCUUGCAUUUUGCACUGCGG ..((((((.((((....))))......--------....((.(((((((.(((((.((((.((....)))))))))))))))))).))..)))))) ( -32.80) >DroYak_CAF1 98164 96 - 1 UGCUGCGGCUGCUGAUCAGCAACCAACUUCCCAAGACAACAAAAUGCAUAGCAUCAUACAAGUGCACGUUGUAGAUGCCUGCAUUUUGCACUGCGG ..((((((.((((....)))).))...............(((((((((..(((((.((((((....).)))))))))).)))))))))....)))) ( -31.10) >consensus UACUGUGGCUGCUGCUAAGCAACCAACUCCCCAAGACAAC__A_UGC___GC__CAUACAA___CAA___GUAGGUACUUGCAUUUUGCAUUGCGG ..((((((.((((....)))).))..............................................((((....))))..........)))) (-10.52 = -10.40 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:42 2006