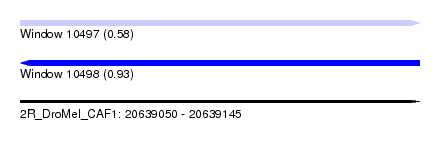
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,639,050 – 20,639,145 |
| Length | 95 |
| Max. P | 0.926696 |

| Location | 20,639,050 – 20,639,145 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 83.74 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -15.02 |
| Energy contribution | -16.19 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20639050 95 + 20766785 CUAUUUGUGGGUGGUCGCUUAUGCCAAGAAAUUU-UAAUUCUUCUUAUAGUCUCUUGGCAACGCGCCCUUAACGCUUU---UUAUGCGUUUUCCGACGU ......((.((.((.(((...((((((((.(((.-(((......))).))).))))))))..))).))..(((((...---....)))))..)).)).. ( -28.20) >DroSec_CAF1 90000 92 + 1 GUAGUUGUGGG---UCGCUUAUGCCAAGAAAUUU-UAAUUCUUCUUAUAGUCUCUUGGCAACGCGCCCUUAACGCUUU---UUAUGCGUUUUCCGACGU ...((((.(((---.(((...((((((((.(((.-(((......))).))).))))))))..))))))..(((((...---....)))))...)))).. ( -30.30) >DroSim_CAF1 90921 92 + 1 GUAGUUGUGGG---UCGCUUAUGCCAAGAAAUUU-UAAUUCUUCUUAUAGUCUCUUGGCAACGCGCCCUUAACGCUUU---UUAUGCAUUUUCCGACGU ...((((.(((---.(((...((((((((.(((.-(((......))).))).))))))))..)))))).))))((...---....))............ ( -27.60) >DroEre_CAF1 128968 92 + 1 GUGGUUGUAGG---CCGCUUAUGCCAAGAAAUUU-AAAUUCUUCUUCUAGUCUCUUGGCAACGCGCCCUCAACGCUUU---UUAUGCGUUUUCCGACGU ...((((.(((---.(((...((((((((.(((.-.((......))..))).))))))))..))).))))))).....---....(((((....))))) ( -26.80) >DroYak_CAF1 93830 92 + 1 GUAGUUGUAGG---CCGCUUAUGCCAAGAAAUUU-AAAUUCUUCUUUUAGUCUCUUGGCAACGCGCCCUUAACGCUUU---UUAUACGUUUUUCGACGA ...((((.(((---.(((...((((((((...((-(((.......)))))..))))))))..))).))))))).....---.....((((....)))). ( -25.50) >DroPer_CAF1 101624 95 + 1 GUGUGUGUG--UGUGUGUGUAUCCCA-GACAUUAAUAUUUGUUCU-UUCGGCUCUUGGCAACGCGCCCUUAACGCAUUUUUUAAUGCGUUUUUUCUCGU ....(((((--(((((.((.....))-.)))).............-....((.....)).))))))....((((((((....))))))))......... ( -19.40) >consensus GUAGUUGUGGG___UCGCUUAUGCCAAGAAAUUU_UAAUUCUUCUUAUAGUCUCUUGGCAACGCGCCCUUAACGCUUU___UUAUGCGUUUUCCGACGU .....(((.((....(((...((((((((.(((...............))).))))))))..(((.......)))..........)))....)).))). (-15.02 = -16.19 + 1.17)


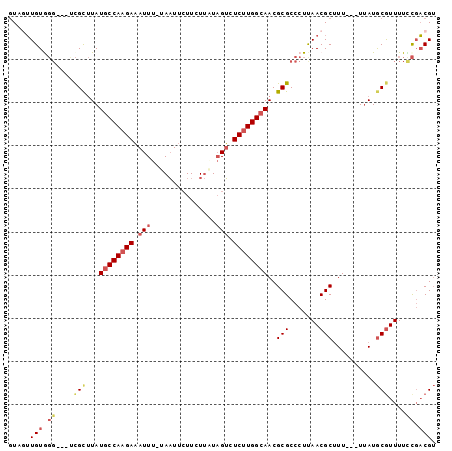
| Location | 20,639,050 – 20,639,145 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 83.74 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -19.09 |
| Energy contribution | -20.07 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.97 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20639050 95 - 20766785 ACGUCGGAAAACGCAUAA---AAAGCGUUAAGGGCGCGUUGCCAAGAGACUAUAAGAAGAAUUA-AAAUUUCUUGGCAUAAGCGACCACCCACAAAUAG ..((.((..(((((....---...)))))..((.(((..((((((((((.(.(((......)))-.).))))))))))...))).)).)).))...... ( -30.50) >DroSec_CAF1 90000 92 - 1 ACGUCGGAAAACGCAUAA---AAAGCGUUAAGGGCGCGUUGCCAAGAGACUAUAAGAAGAAUUA-AAAUUUCUUGGCAUAAGCGA---CCCACAACUAC .........(((((....---...)))))..((((((..((((((((((.(.(((......)))-.).))))))))))...))).---)))........ ( -30.40) >DroSim_CAF1 90921 92 - 1 ACGUCGGAAAAUGCAUAA---AAAGCGUUAAGGGCGCGUUGCCAAGAGACUAUAAGAAGAAUUA-AAAUUUCUUGGCAUAAGCGA---CCCACAACUAC .........(((((....---...)))))..((((((..((((((((((.(.(((......)))-.).))))))))))...))).---)))........ ( -28.60) >DroEre_CAF1 128968 92 - 1 ACGUCGGAAAACGCAUAA---AAAGCGUUGAGGGCGCGUUGCCAAGAGACUAGAAGAAGAAUUU-AAAUUUCUUGGCAUAAGCGG---CCUACAACCAC .....((..(((((....---...))))).(((.(((..((((((((((.(.(((......)))-.).))))))))))...))).---)))....)).. ( -32.70) >DroYak_CAF1 93830 92 - 1 UCGUCGAAAAACGUAUAA---AAAGCGUUAAGGGCGCGUUGCCAAGAGACUAAAAGAAGAAUUU-AAAUUUCUUGGCAUAAGCGG---CCUACAACUAC .........(((((....---...))))).(((.(((..((((((((((.(.(((......)))-.).))))))))))...))).---)))........ ( -28.00) >DroPer_CAF1 101624 95 - 1 ACGAGAAAAAACGCAUUAAAAAAUGCGUUAAGGGCGCGUUGCCAAGAGCCGAA-AGAACAAAUAUUAAUGUC-UGGGAUACACACACA--CACACACAC .(.(((...((((((((....))))))))...(((.(........).)))...-................))-).)............--......... ( -16.70) >consensus ACGUCGGAAAACGCAUAA___AAAGCGUUAAGGGCGCGUUGCCAAGAGACUAUAAGAAGAAUUA_AAAUUUCUUGGCAUAAGCGA___CCCACAACUAC .........(((((..........)))))..((((((..((((((((((...(((......)))....))))))))))...)))....)))........ (-19.09 = -20.07 + 0.97)

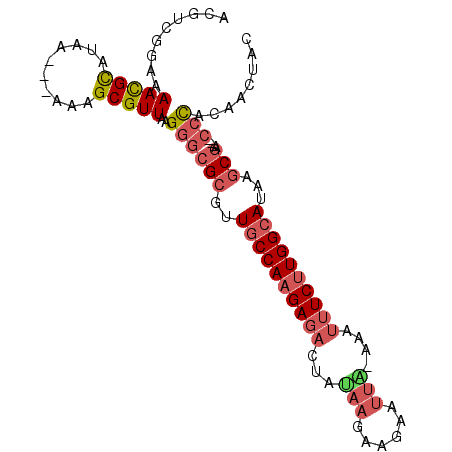
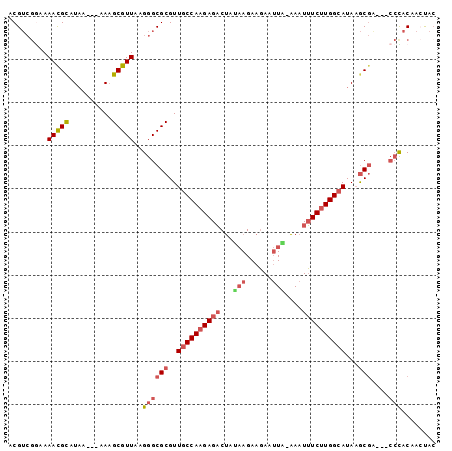
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:41 2006