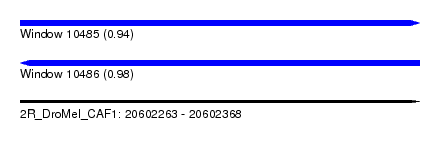
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,602,263 – 20,602,368 |
| Length | 105 |
| Max. P | 0.983325 |

| Location | 20,602,263 – 20,602,368 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.13 |
| Mean single sequence MFE | -51.47 |
| Consensus MFE | -31.35 |
| Energy contribution | -31.88 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
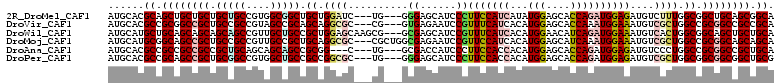
>2R_DroMel_CAF1 20602263 105 + 20766785 UGCCGCUGCAGCCGCCAAAGACAUCUCCAUCUGGUGCUCCAUAUGAUGGAAGGGAUGCUCCC---CA---GAUCCAGCAGCCGCCACGGCAGCAGCAGCAGCUGCGUGCAU (((((..((.((.((..........(((((((((....)))...)))))).(((......))---).---......)).)).))..)))))(((((....)))))...... ( -38.80) >DroVir_CAF1 101854 105 + 1 UGCGGCGGCCGCGGCCAGCGACAUUUCCAUUUGGUGCUCCAUGUGAUGAAACGGAUUCUCAC---CG---GCGCCUGCUGCGGCUACGGCGGCAGCGGCCGCGGCGUGCAU ....((((((((((((................((((((....((((.(((.....)))))))---.)---))))).(((((.((....)).)))))))))))))).))).. ( -56.20) >DroWil_CAF1 97440 108 + 1 UGCAGCAGCUGCCGCCAGUGACAUUUCCAUCUGAUGUUCCAUGUGAUGAAACGGAUGCUCGC---CGCUUGCUCCAGCGGCAGCAACGGCUGCUGCUGCUGCAGCAUGCAU .((((((((.((((((.(..(((((.......)))))..)..((......)))).((((.((---((((......)))))))))).)))).))))))))(((.....))). ( -48.80) >DroMoj_CAF1 113407 108 + 1 UGCUGCUGCCGCGGCCAGCGACAUUUCCAUUUGAUGCUCCAUGUGAUGGAACGGAUUCUCGCCAGCG---GCGCCUGCAGCGGCAACGGCGGCAGCGGCUGCCGCAUGCAU ((((((((((((.((..((((....(((.(((.((((.....)).)).))).)))...))))..)).---)))...))))))))).(((((((....)))))))....... ( -48.50) >DroAna_CAF1 58425 102 + 1 UGCAGCGGCCGCGGCCAGGGACAUCUCCAUCUGGUGCUCCAUGUGGUGGAAGGGAUGGUCGC---CA---G---CCGCGGCUGCUGCUGCAGCGGCGGCGGCGGCGUGCAU ((((((.((((((((((....(.(((((((.(((....))).)))).))).)...)))))))---..---(---((((.(((((....))))).)))))))).)).)))). ( -60.70) >DroPer_CAF1 64712 105 + 1 CGCAGCCGCCGCCGCCAGCGACAUCUCCAUCUGGUGCUCCAUGUGGUGGAAGGGAUGCUCCC---CA---GCGCCGGCGGCAGCCACGGCCGCAGCGGCUGCGGCGUGCAU (((((((((.((((((.(((.(.(((((((.(((....))).)))).))).(((......))---).---)))).)))))).((....))....)))))))))........ ( -55.80) >consensus UGCAGCGGCCGCCGCCAGCGACAUCUCCAUCUGGUGCUCCAUGUGAUGGAACGGAUGCUCGC___CA___GCGCCAGCGGCAGCAACGGCAGCAGCGGCUGCGGCGUGCAU .(((...(((((.(((...((((((((((((..(((...)))..))))))...)))).))................((.((.((....)).)).))))).))))).))).. (-31.35 = -31.88 + 0.53)

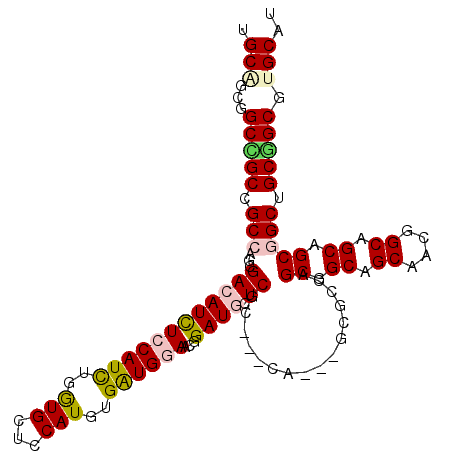

| Location | 20,602,263 – 20,602,368 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.13 |
| Mean single sequence MFE | -53.90 |
| Consensus MFE | -32.25 |
| Energy contribution | -33.28 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.36 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
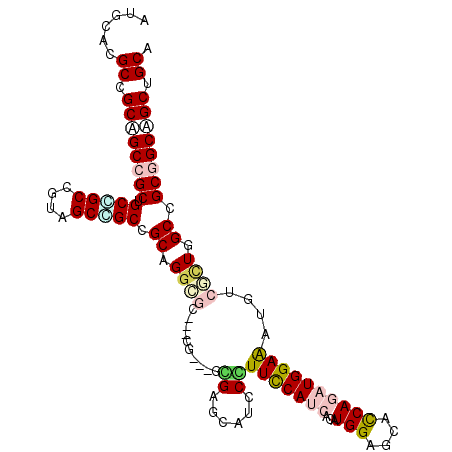
>2R_DroMel_CAF1 20602263 105 - 20766785 AUGCACGCAGCUGCUGCUGCUGCCGUGGCGGCUGCUGGAUC---UG---GGGAGCAUCCCUUCCAUCAUAUGGAGCACCAGAUGGAGAUGUCUUUGGCGGCUGCAGCGGCA ......(((((....))))).(((((.((((((((..((..---.(---(((.....))))((((((...(((....)))))))))......))..)))))))).))))). ( -52.70) >DroVir_CAF1 101854 105 - 1 AUGCACGCCGCGGCCGCUGCCGCCGUAGCCGCAGCAGGCGC---CG---GUGAGAAUCCGUUUCAUCACAUGGAGCACCAAAUGGAAAUGUCGCUGGCCGCGGCCGCCGCA ......((.(((((((((((.((....)).)))))..((((---((---((((.(....(((((((...))))))).((....))...).))))))).))))))))).)). ( -52.20) >DroWil_CAF1 97440 108 - 1 AUGCAUGCUGCAGCAGCAGCAGCCGUUGCUGCCGCUGGAGCAAGCG---GCGAGCAUCCGUUUCAUCACAUGGAACAUCAGAUGGAAAUGUCACUGGCGGCAGCUGCUGCA .(((.....)))((((((((.(((((((((((((((......))))---)).))))...(((((((...)))))))..(((.(((.....))))))))))).)))))))). ( -52.30) >DroMoj_CAF1 113407 108 - 1 AUGCAUGCGGCAGCCGCUGCCGCCGUUGCCGCUGCAGGCGC---CGCUGGCGAGAAUCCGUUCCAUCACAUGGAGCAUCAAAUGGAAAUGUCGCUGGCCGCGGCAGCAGCA ..(((.(((((((...)))))))...))).(((((.(.(((---.((..((((.(.((((((((((...))))))).......)))..).))))..)).))).).))))). ( -53.21) >DroAna_CAF1 58425 102 - 1 AUGCACGCCGCCGCCGCCGCUGCAGCAGCAGCCGCGG---C---UG---GCGACCAUCCCUUCCACCACAUGGAGCACCAGAUGGAGAUGUCCCUGGCCGCGGCCGCUGCA .((((.((.((....)).))))))(((((.(((((((---(---..---(.(((.(((...((((.....(((....)))..)))))))))).)..)))))))).))))). ( -53.40) >DroPer_CAF1 64712 105 - 1 AUGCACGCCGCAGCCGCUGCGGCCGUGGCUGCCGCCGGCGC---UG---GGGAGCAUCCCUUCCACCACAUGGAGCACCAGAUGGAGAUGUCGCUGGCGGCGGCGGCUGCG ..(((.(((((((...))))))).((.((((((((((((((---.(---(((.....))))....(((..(((....)))..)))....).))))))))))))).))))). ( -59.60) >consensus AUGCACGCCGCAGCCGCUGCCGCCGUAGCCGCCGCAGGCGC___CG___GCGAGCAUCCCUUCCAUCACAUGGAGCACCAGAUGGAAAUGUCGCUGGCCGCGGCAGCUGCA ......((.((((((((.(((((....))))).((.((((..........((......))(((((((...(((....))))))))))....)))).)).)))))))).)). (-32.25 = -33.28 + 1.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:29 2006