| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,595,246 – 20,595,348 |
| Length | 102 |
| Max. P | 0.766378 |

| Location | 20,595,246 – 20,595,348 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.68 |
| Mean single sequence MFE | -44.63 |
| Consensus MFE | -32.29 |
| Energy contribution | -32.27 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
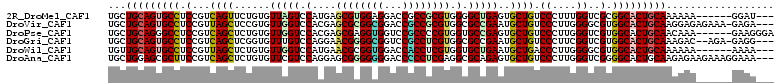
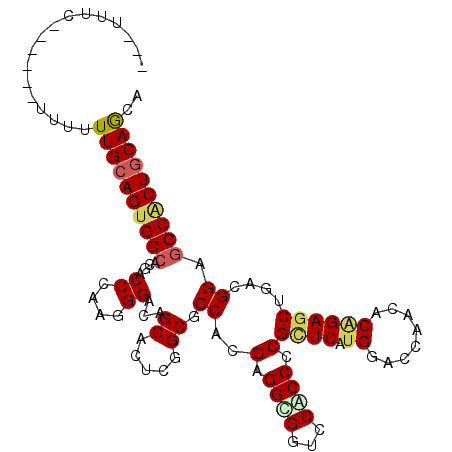
>2R_DroMel_CAF1 20595246 102 + 20766785 UGCUGCAGUGCCUCCGUCAGUUCUGUGUUAGUCCAUGAGCGUGGAGGACCGCCGCGUGGGGCUGAGUGCUGUCCCUUUGGUCGCGGCACUGCAAAAAA------GGAU--- ...((((((.((((((...((((.(((......))))))).))))))...((((((.(((((........)))))......)))))))))))).....------....--- ( -42.50) >DroVir_CAF1 92935 107 + 1 UGCUGCAGUGCCUCCGUUAGCUCCGUGUUGGUCCACGAGCGCGGCGGACCGCCGCGUGGCGCCGAAUGCUGUCCCUUGGGGCGUGGCACUGCAAGGAGAGAAA-GAGA--- ...(((((((((.(....(((......((((((((((.(.((((....))))).))))).)))))..)))((((....))))).)))))))))..........-....--- ( -47.30) >DroPse_CAF1 48895 105 + 1 UGCUGCAGGGCCUCCGUCAGCUCUGUGUUGGUCCACGAGCGAGGUGGUCCGCCCCGUGGUGCCGAGUGCUGUCCCUUGGGUCGUGGCACUGCAACAAA------GAAGGGA ....((.(((((.((.((.((((.(((......))))))))))).)))))))(((.((.(((..(((((..(((....))..)..)))))))).))..------...))). ( -41.60) >DroGri_CAF1 97354 105 + 1 UGCUGCAGUGCCUCCGUCAGCUCGGUGUUUGUCCAGGAACGGGGCGGUCCGCCUCGUGGCGCCGAAUGCUGUCCCUUCGGUCGUGGCACUGCAAAGAC--AGA-GAGG--- ...(((((((((.(((.((((((((((((.........((((((((...))))))))))))))))..))))......)))....))))))))).....--...-....--- ( -46.70) >DroWil_CAF1 91116 102 + 1 UGUUGCAGUGCCUCCGUUAGCUCUGUGUUGGUCCAUGAACGCGGUGGACCACCUCGUGGUGCUGAAUGCUGACCCUUGGGGCGUGGCACUGCAAAAAA------AAAA--- ..((((((((((....(((((.(..((.((((((((.......))))))))...))..).)))))((((...((....))))))))))))))))....------....--- ( -44.40) >DroAna_CAF1 53317 108 + 1 UGCUGGAGCGCUUCCGUCAGCUCUGUGUUCGUCCAGGAGCGGGGGGGACCCCCUCGAGGCGCAGAGUGCUGUCCCUUGGGUCGGGGCACUGCAAGAGAAGAAAGGAAA--- .((....))..((((.((..((((......(.((..(((.((((....)))))))..)))((((.((.(((.((....)).))).)).)))).))))..))..)))).--- ( -45.30) >consensus UGCUGCAGUGCCUCCGUCAGCUCUGUGUUGGUCCACGAGCGCGGCGGACCGCCUCGUGGCGCCGAAUGCUGUCCCUUGGGUCGUGGCACUGCAAAAAA______GAAA___ ...(((((((((.(...((((......(((((.(....((((((((...)))))))).).)))))..)))).((....))..).))))))))).................. (-32.29 = -32.27 + -0.03)
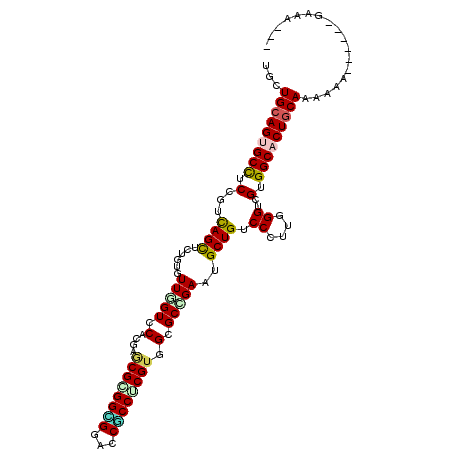
| Location | 20,595,246 – 20,595,348 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.68 |
| Mean single sequence MFE | -39.50 |
| Consensus MFE | -27.75 |
| Energy contribution | -28.12 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20595246 102 - 20766785 ---AUCC------UUUUUUGCAGUGCCGCGACCAAAGGGACAGCACUCAGCCCCACGCGGCGGUCCUCCACGCUCAUGGACUAACACAGAACUGACGGAGGCACUGCAGCA ---....------....((((((((((..((((...(((......))).(((......))))))).....((.(((.(..((.....))..))))))..)))))))))).. ( -33.20) >DroVir_CAF1 92935 107 - 1 ---UCUC-UUUCUCUCCUUGCAGUGCCACGCCCCAAGGGACAGCAUUCGGCGCCACGCGGCGGUCCGCCGCGCUCGUGGACCAACACGGAGCUAACGGAGGCACUGCAGCA ---....-.........((((((((((.(((((....)((......))))))((..((((((...))))))(((((((......))).))))....)).)))))))))).. ( -45.50) >DroPse_CAF1 48895 105 - 1 UCCCUUC------UUUGUUGCAGUGCCACGACCCAAGGGACAGCACUCGGCACCACGGGGCGGACCACCUCGCUCGUGGACCAACACAGAGCUGACGGAGGCCCUGCAGCA .......------..((((((((.(((.........((..(.((.((((......)))))))..)).(((((((((((......))).)))).)).)).))).)))))))) ( -42.40) >DroGri_CAF1 97354 105 - 1 ---CCUC-UCU--GUCUUUGCAGUGCCACGACCGAAGGGACAGCAUUCGGCGCCACGAGGCGGACCGCCCCGUUCCUGGACAAACACCGAGCUGACGGAGGCACUGCAGCA ---....-...--....((((((((((.((.((....)).((((..((((.((((.(..((((......))))..)))).....).)))))))).))..)))))))))).. ( -42.90) >DroWil_CAF1 91116 102 - 1 ---UUUU------UUUUUUGCAGUGCCACGCCCCAAGGGUCAGCAUUCAGCACCACGAGGUGGUCCACCGCGUUCAUGGACCAACACAGAGCUAACGGAGGCACUGCAACA ---....------....((((((((((..((((...))))........((((((....)))((((((.........))))))........)))......)))))))))).. ( -36.30) >DroAna_CAF1 53317 108 - 1 ---UUUCCUUUCUUCUCUUGCAGUGCCCCGACCCAAGGGACAGCACUCUGCGCCUCGAGGGGGUCCCCCCCGCUCCUGGACGAACACAGAGCUGACGGAAGCGCUCCAGCA ---...............(((((((((((.......)))...)))))..((((.((..(((((...)))))((((.((........)))))).....)).))))....))) ( -36.70) >consensus ___UUUC______UUUUUUGCAGUGCCACGACCCAAGGGACAGCACUCGGCGCCACGAGGCGGUCCACCCCGCUCAUGGACCAACACAGAGCUGACGGAGGCACUGCAGCA .................((((((((((....((....))...((.....)).((..(.((((...)))).)((((.((........))))))....)).)))))))))).. (-27.75 = -28.12 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:26 2006