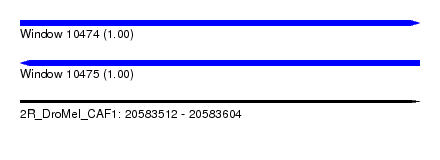
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,583,512 – 20,583,604 |
| Length | 92 |
| Max. P | 0.999720 |

| Location | 20,583,512 – 20,583,604 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 74.06 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -16.18 |
| Energy contribution | -15.82 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.59 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20583512 92 + 20766785 AUAGAAAAAAUACCUGCCUUACCGACG-UAUUGCGGUAAAGCAGUUU-UUCUUGGCCUGCCUGUAGUAAUGGUUUAGGCAGCACUCAUGACCAA ..(((((((....((((.((((((...-.....)))))).)))))))-))))..((.((((((...........))))))))............ ( -25.90) >DroSec_CAF1 35193 93 + 1 AUAGGUAAAAUACCUGCCUUACCGACG-UAUUGCGGUAAAGCAGUUUUUUCUUGGCCUGCCUCUACUAAUGAGUUAGGCAGAAUAUCAGACAAA ...((((......((((.((((((...-.....)))))).))))....((((..(((((.(((.......))).)))))))))))))....... ( -27.00) >DroSim_CAF1 35262 93 + 1 AUAGGUAAAAUACCUGCCUUACCGACG-AAUUGCGGUUAAGCAGUUUUUUCUUGUCCUGCCUCUAGUAAUGUGUUAGGCAGGGUAACAGACAAA .((((((...))))))..(((((((((-((((((......)))))......)))))((((((.((....))....)))))))))))........ ( -24.10) >DroEre_CAF1 62722 89 + 1 -----AAAAGGAUCUGCUUUACCGACGCAAGUGUGGUAAAGCAGUUUUUCCUUGGCCAGUCACUAGUGCUGCAUUCGGCUCUAAUGUUGACCAA -----..(((((.(((((((((((((....)).)))))))))))....)))))((.(((.((.(((.((((....)))).))).))))).)).. ( -37.50) >DroYak_CAF1 37078 90 + 1 A----UAAAACCCCUGCUUUACCGACCUUAUUGUGGUAAAGCAGUUUUUUCUUGGACUGUCACUAGUGGUGCAUUAGACAAAAGUGAUGAACAA .----........((((((((((.((......))))))))))))........((..(.((((((..((..........))..)))))))..)). ( -22.50) >consensus AUAG_UAAAAUACCUGCCUUACCGACG_UAUUGCGGUAAAGCAGUUUUUUCUUGGCCUGCCUCUAGUAAUGCAUUAGGCAGAAUUGAUGACCAA .............((((.((((((.(......))))))).))))............(((((...............)))))............. (-16.18 = -15.82 + -0.36)

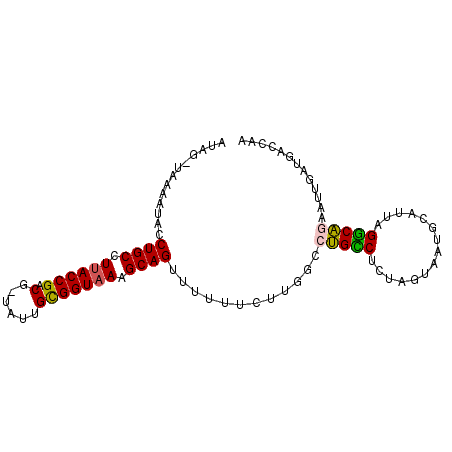
| Location | 20,583,512 – 20,583,604 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 74.06 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -20.78 |
| Energy contribution | -20.18 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.73 |
| SVM decision value | 3.94 |
| SVM RNA-class probability | 0.999720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20583512 92 - 20766785 UUGGUCAUGAGUGCUGCCUAAACCAUUACUACAGGCAGGCCAAGAA-AAACUGCUUUACCGCAAUA-CGUCGGUAAGGCAGGUAUUUUUUCUAU .((((........((((((.............))))))))))((((-(((((((((((((((....-.).)))))))))))....))))))).. ( -30.12) >DroSec_CAF1 35193 93 - 1 UUUGUCUGAUAUUCUGCCUAACUCAUUAGUAGAGGCAGGCCAAGAAAAAACUGCUUUACCGCAAUA-CGUCGGUAAGGCAGGUAUUUUACCUAU ...((..((((((((((((..(((.......)))..))))..))).....((((((((((((....-.).))))))))))))))))..)).... ( -30.10) >DroSim_CAF1 35262 93 - 1 UUUGUCUGUUACCCUGCCUAACACAUUACUAGAGGCAGGACAAGAAAAAACUGCUUAACCGCAAUU-CGUCGGUAAGGCAGGUAUUUUACCUAU ....(((.....(((((((.............)))))))...))).....((((((.(((((....-.).)))).))))))............. ( -26.02) >DroEre_CAF1 62722 89 - 1 UUGGUCAACAUUAGAGCCGAAUGCAGCACUAGUGACUGGCCAAGGAAAAACUGCUUUACCACACUUGCGUCGGUAAAGCAGAUCCUUUU----- (((((((.((((((.((........)).))))))..)))))))(((....((((((((((((......)).)))))))))).)))....----- ( -33.60) >DroYak_CAF1 37078 90 - 1 UUGUUCAUCACUUUUGUCUAAUGCACCACUAGUGACAGUCCAAGAAAAAACUGCUUUACCACAAUAAGGUCGGUAAAGCAGGGGUUUUA----U (((..(.(((((..((..........))..)))))..)..)))((((...((((((((((((......)).))))))))))...)))).----. ( -22.80) >consensus UUGGUCAGCAAUUCUGCCUAACGCAUUACUAGAGGCAGGCCAAGAAAAAACUGCUUUACCGCAAUA_CGUCGGUAAGGCAGGUAUUUUA_CUAU .............((((((.............))))))............((((((((((((......)).))))))))))............. (-20.78 = -20.18 + -0.60)

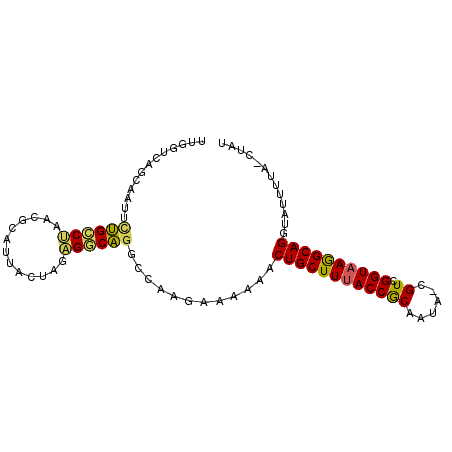
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:19 2006