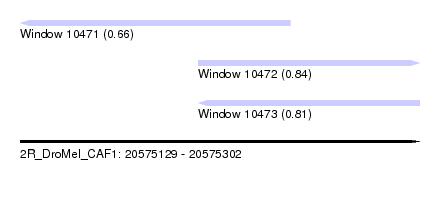
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,575,129 – 20,575,302 |
| Length | 173 |
| Max. P | 0.837289 |

| Location | 20,575,129 – 20,575,246 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.49 |
| Mean single sequence MFE | -25.56 |
| Consensus MFE | -15.40 |
| Energy contribution | -15.18 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20575129 117 - 20766785 AUUGAACACGGCACGUAUACUAACUACGUGCGCGCGUUUUGUAAAAUAACAUUCAAUUUGUGCAACCUGAUCAAGCCAUUGCAGCAGGACCAGAUUAUUGAUCGCCCUUCGACGAUC--- ..........(((((((.......)))))))(((((..(((............)))..)))))..((((..(((....)))...))))....(((..((((.......))))..)))--- ( -28.30) >DroVir_CAF1 40545 106 - 1 AUCGCAUAUUGCACGG---CUAGCUACGUGUGCGCAUUUUGCAAAAUAACAUUCAAUUUGUGCAACCUGAUCAAGCAAUUUCAUCAAC----------CGAUCAG-GUCCAACGAUCUCU ((((......(((((.---.......)))))(((((..(((.((.......)))))..))))).((((((((................----------.))))))-))....)))).... ( -25.13) >DroPse_CAF1 29343 103 - 1 GUGGCAUACUGCACGUAUACUUGCUACGUGCGCGCAUUUUGCAAAAUAACAUUCAAUUUGUGCAACCUGAUCAAGCAAUUGCAUCA--------------ACCGCUGAUCGAUGAUC--- ((((......(((((((.......)))))))(((((..(((.((.......)))))..)))))....(((((((....))).))))--------------.)))).((((...))))--- ( -26.50) >DroGri_CAF1 43077 103 - 1 AUCGCAUAUUGCACGG---CUAGCUACGUGUGCGCAUUUUGCAAAAUAACAUUCAAUUUGUGCAACCUGAUCAAGCAAUUUCAUCAGG----------CAUUCAA----CCUCGAUCUAC ((((......(((((.---.......)))))(((((..(((.((.......)))))..)))))..((((((.((....))..))))))----------.......----...)))).... ( -21.20) >DroMoj_CAF1 44442 104 - 1 AUCGCAUAUUGCACGG---CUAGGUACGUGUGCGCAUUUUGCAAAAUAACAUUCAAUUUGUGCAACCUGAUCAAGCAAUUGCAACAAC----------CGAUCGC-GUCCAAUGAUC--U ...(((.(((((..(.---.(((((.....((((((..(((.((.......)))))..)))))))))))..)..))))))))......----------.(((((.-......)))))--. ( -25.70) >DroPer_CAF1 29279 103 - 1 GUGGCAUACUGCACGUAUACUUGCUACGUGCGCGCAUUUUGCAAAAUAACAUUCAAUUUGUGCAACCUGAUCAAGCAAUUGCAUCA--------------ACCGCUGAUCGAUGAUC--- ((((......(((((((.......)))))))(((((..(((.((.......)))))..)))))....(((((((....))).))))--------------.)))).((((...))))--- ( -26.50) >consensus AUCGCAUACUGCACGG___CUAGCUACGUGCGCGCAUUUUGCAAAAUAACAUUCAAUUUGUGCAACCUGAUCAAGCAAUUGCAUCA____________CGAUCGC_GUUCGACGAUC___ ...((.....(((((...........)))))(((((..(((............)))..)))))...........))............................................ (-15.40 = -15.18 + -0.22)


| Location | 20,575,206 – 20,575,302 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 79.25 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -17.63 |
| Energy contribution | -18.63 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20575206 96 + 20766785 AAAACGCGCGCACGUAGUUAGUAUACGUGCCGUGUUCAAU-----UGCAGAAGACGACUAAGUCCUGACCCAGUAGAUUGGGGGGGGAAGGGUGGGUCGGG ..((((((.(((((((.......)))))))))))))....-----.........(((((...((((..(((((....)))))..))))......))))).. ( -37.70) >DroPse_CAF1 29406 87 + 1 AAAAUGCGCGCACGUAGCAAGUAUACGUGCAGUAUGCCACAGCGUGGCAGCAGAAU--AAAGUUACCUUUCUGGGGAUAGAG------------AGAAGGG ....(((..(((((((.......)))))))....((((((...)))))))))....--........(((((((....)))))------------))..... ( -28.40) >DroSec_CAF1 26712 85 + 1 AAAAUGCGCGCACGUAGUUAGUAUACGUGCCGUGUUCAAU-----UGCAGAAGACGACUAAGUCCUGACCCAGGAGAUGGGG-----------GGAUCGGG .........(((((((.......)))))))(((.(((...-----....))).))).((..(((((..((((.....)))))-----------))))..)) ( -26.60) >DroSim_CAF1 26841 85 + 1 AAAAUGCGCGCACGUAGUUAGUAUACGUGCCGUGUUCAAU-----UGCAGAAGACGACUAAGUCCUGACCCAGGAGAUGGGG-----------GGGUCGGG ..((((((.(((((((.......)))))))))))))....-----.......(((......)))(((((((...........-----------))))))). ( -28.60) >DroEre_CAF1 27248 84 + 1 AAAAUGCGCGCACGUAGUUAGUAGACGUGCCGUGUUCAAU-----UGCAGAAGACGACCAAGUCCUGACCCAGGAGAAGGG------------GAAUCGGG ..((((((.((((((.........))))))))))))....-----.......(((......)))(((((((........))------------)..)))). ( -21.80) >DroYak_CAF1 27686 84 + 1 AAAAUGCGCGCACGUAGUUAGUAUACGUGCCGUGUUCAAU-----UGCAGAAGACGACUAAGUCCUGACCCAGCAGAUGGG------------GGAUUGGG ..((((((.(((((((.......)))))))))))))((((-----(.(((..(((......)))))).((((.....))))------------.))))).. ( -28.00) >consensus AAAAUGCGCGCACGUAGUUAGUAUACGUGCCGUGUUCAAU_____UGCAGAAGACGACUAAGUCCUGACCCAGGAGAUGGGG___________GGAUCGGG ..((((((.(((((((.......)))))))))))))...........(((..(((......)))))).((((.....)))).................... (-17.63 = -18.63 + 1.00)

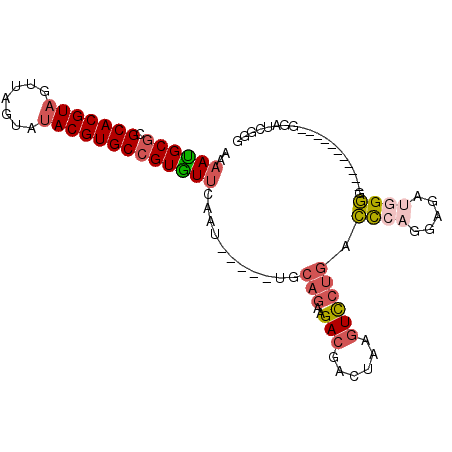
| Location | 20,575,206 – 20,575,302 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 79.25 |
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -15.18 |
| Energy contribution | -14.77 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20575206 96 - 20766785 CCCGACCCACCCUUCCCCCCCCAAUCUACUGGGUCAGGACUUAGUCGUCUUCUGCA-----AUUGAACACGGCACGUAUACUAACUACGUGCGCGCGUUUU ...((((((....................))))))(((((......))))).....-----...((((.(((((((((.......))))))).)).)))). ( -25.95) >DroPse_CAF1 29406 87 - 1 CCCUUCU------------CUCUAUCCCCAGAAAGGUAACUUU--AUUCUGCUGCCACGCUGUGGCAUACUGCACGUAUACUUGCUACGUGCGCGCAUUUU .((((.(------------((........))))))).......--....(((((((((...))))))....(((((((.......)))))))..))).... ( -23.20) >DroSec_CAF1 26712 85 - 1 CCCGAUCC-----------CCCCAUCUCCUGGGUCAGGACUUAGUCGUCUUCUGCA-----AUUGAACACGGCACGUAUACUAACUACGUGCGCGCAUUUU ((.(((((-----------...........))))).)).......(((.(((....-----...))).)))(((((((.......)))))))......... ( -20.20) >DroSim_CAF1 26841 85 - 1 CCCGACCC-----------CCCCAUCUCCUGGGUCAGGACUUAGUCGUCUUCUGCA-----AUUGAACACGGCACGUAUACUAACUACGUGCGCGCAUUUU ((.(((((-----------...........))))).)).......(((.(((....-----...))).)))(((((((.......)))))))......... ( -22.80) >DroEre_CAF1 27248 84 - 1 CCCGAUUC------------CCCUUCUCCUGGGUCAGGACUUGGUCGUCUUCUGCA-----AUUGAACACGGCACGUCUACUAACUACGUGCGCGCAUUUU .((((.((------------((((......)))...))).)))).(((.(((....-----...))).)))((((((.........))))))......... ( -19.80) >DroYak_CAF1 27686 84 - 1 CCCAAUCC------------CCCAUCUGCUGGGUCAGGACUUAGUCGUCUUCUGCA-----AUUGAACACGGCACGUAUACUAACUACGUGCGCGCAUUUU .....(((------------((((.....))))...)))......(((.(((....-----...))).)))(((((((.......)))))))......... ( -19.60) >consensus CCCGACCC___________CCCCAUCUCCUGGGUCAGGACUUAGUCGUCUUCUGCA_____AUUGAACACGGCACGUAUACUAACUACGUGCGCGCAUUUU ((.(((((......................))))).)).......(((.(((............))).)))(((((((.......)))))))......... (-15.18 = -14.77 + -0.41)

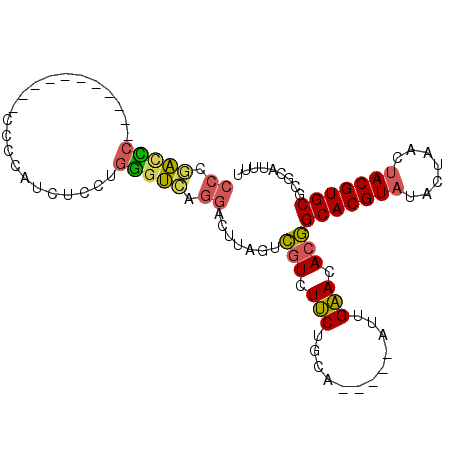
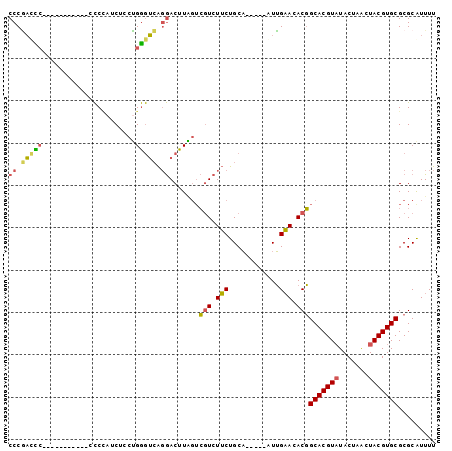
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:17 2006