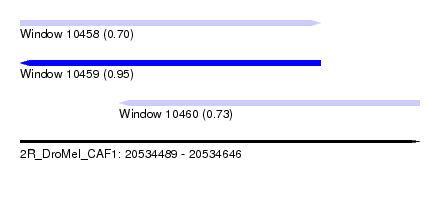
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,534,489 – 20,534,646 |
| Length | 157 |
| Max. P | 0.948970 |

| Location | 20,534,489 – 20,534,607 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -18.88 |
| Energy contribution | -20.44 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20534489 118 + 20766785 AAAGUUACUUGGCCCCGACGAGGGGAGUGUCAUCC-CUCGUCGAGUUUGUCUUCUGAAACA-UAUCACAAUAACACAUCAUUUGUCAAGGCAACAAAUGAUGUUGCAUGUAACCUUAUCA ...(((((...((..((((((((((.......)))-))))))).((((........)))).-............(((((((((((.......))))))))))).))..)))))....... ( -35.90) >DroSim_CAF1 36374 118 + 1 ACAGUUACUUGGCCCCGGCGAGGGCAGUGUCAUCCCCUCGUCGAGUUUCGCUUC-AAAACA-UAUCACAAUAACACAUCAUUUGUCAAGGCAACAAAUGAUGUUGCAUGUAACCUUAUCA ...(((((...((..(((((((((.(......).)))))))))...........-......-............(((((((((((.......))))))))))).))..)))))....... ( -33.90) >DroYak_CAF1 37843 119 + 1 AAAGUUACUUGGCCCCGGCGAGCGUAAUGUCAUCCCCUCGUUGAGUUUGGCUUCUGAAACA-UAUCACAAUAACACAUCAUUUGUCAAGGCAACAAAUGAUGUUGCAUGUAACCUUAUCA ...(((((.(((((((((((((.(..((...))..)))))))).)...)))..........-............(((((((((((.......)))))))))))..)).)))))....... ( -26.30) >DroAna_CAF1 44351 100 + 1 AAAGUUGCUUG-CCCCGGCUGGG------UC-------------GUUCGGCUUUUGAAACACUAUCACAAUAACACAUCAUUUGUCAGGCAAACAAACGAUGUUGCAUGUAACCUUAUCA ...(((((.((-(...((((((.------..-------------..))))))......................(((((.(((((.......))))).))))).))).)))))....... ( -20.00) >consensus AAAGUUACUUGGCCCCGGCGAGGG_AGUGUCAUCC_CUCGUCGAGUUUGGCUUCUGAAACA_UAUCACAAUAACACAUCAUUUGUCAAGGCAACAAAUGAUGUUGCAUGUAACCUUAUCA ...(((((.((....(((((((..............)))))))...............................(((((((((((.......)))))))))))..)).)))))....... (-18.88 = -20.44 + 1.56)

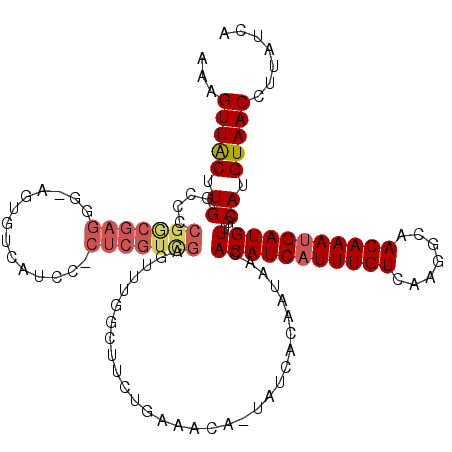
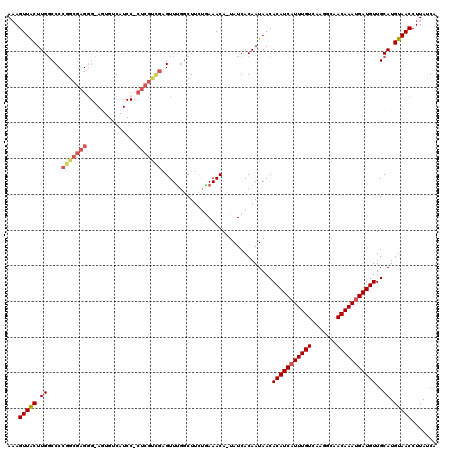
| Location | 20,534,489 – 20,534,607 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -36.27 |
| Consensus MFE | -23.25 |
| Energy contribution | -25.12 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20534489 118 - 20766785 UGAUAAGGUUACAUGCAACAUCAUUUGUUGCCUUGACAAAUGAUGUGUUAUUGUGAUA-UGUUUCAGAAGACAAACUCGACGAG-GGAUGACACUCCCCUCGUCGGGGCCAAGUAACUUU ....((((((((..((.((((((((((((.....))))))))))))(((.((.(((..-....))).)))))...(((((((((-((.........)))))))))))))...)))))))) ( -46.20) >DroSim_CAF1 36374 118 - 1 UGAUAAGGUUACAUGCAACAUCAUUUGUUGCCUUGACAAAUGAUGUGUUAUUGUGAUA-UGUUUU-GAAGCGAAACUCGACGAGGGGAUGACACUGCCCUCGCCGGGGCCAAGUAACUGU ......((((((..((.((((((((((((.....))))))))))))....((((..((-.....)-)..))))..((((.((((((.(......).)))))).))))))...)))))).. ( -38.40) >DroYak_CAF1 37843 119 - 1 UGAUAAGGUUACAUGCAACAUCAUUUGUUGCCUUGACAAAUGAUGUGUUAUUGUGAUA-UGUUUCAGAAGCCAAACUCAACGAGGGGAUGACAUUACGCUCGCCGGGGCCAAGUAACUUU ....((((((((..((.((((((((((((.....))))))))))))....((.(((..-....))).))))....(((..((((.(..........).))))..))).....)))))))) ( -33.20) >DroAna_CAF1 44351 100 - 1 UGAUAAGGUUACAUGCAACAUCGUUUGUUUGCCUGACAAAUGAUGUGUUAUUGUGAUAGUGUUUCAAAAGCCGAAC-------------GA------CCCAGCCGGGG-CAAGCAACUUU .((((..((((((.(((.(((((((((((.....))))))))))))))...))))))..))))......(((...(-------------(.------......)).))-).......... ( -27.30) >consensus UGAUAAGGUUACAUGCAACAUCAUUUGUUGCCUUGACAAAUGAUGUGUUAUUGUGAUA_UGUUUCAGAAGCCAAACUCGACGAG_GGAUGACACU_CCCUCGCCGGGGCCAAGUAACUUU .((((..((((((.(((.(((((((((((.....))))))))))))))...))))))..))))............((((.((((.(..........).)))).))))............. (-23.25 = -25.12 + 1.88)

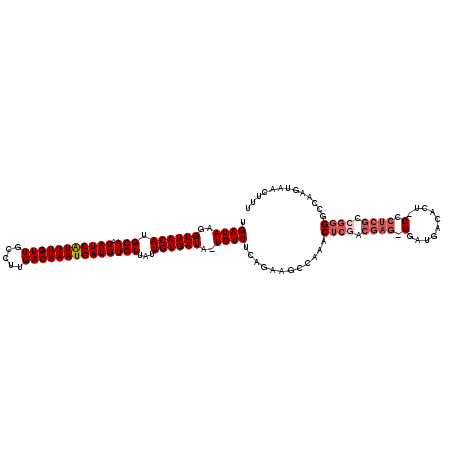
| Location | 20,534,528 – 20,534,646 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.25 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -25.58 |
| Energy contribution | -25.45 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20534528 118 - 20766785 UGGCAA-ACCGUAACGUAACCGUCUGCAACGGAUAUGAGAUGAUAAGGUUACAUGCAACAUCAUUUGUUGCCUUGACAAAUGAUGUGUUAUUGUGAUA-UGUUUCAGAAGACAAACUCGA ......-..(((..((((.((((.....)))).))))..))).....((((((.(((.(((((((((((.....))))))))))))))...)))))).-((((......))))....... ( -30.20) >DroSim_CAF1 36414 117 - 1 UGCCAA-ACCGUAACGUAACCGUCUGCAGCGGAUAUGAGAUGAUAAGGUUACAUGCAACAUCAUUUGUUGCCUUGACAAAUGAUGUGUUAUUGUGAUA-UGUUUU-GAAGCGAAACUCGA .(((((-(.(((..((((.((((.....)))).))))..........((((((.(((.(((((((((((.....))))))))))))))...)))))))-)).)))-)..))......... ( -31.30) >DroYak_CAF1 37883 118 - 1 UGCCAA-ACCGUAACGUAAGCGUCUGCAGCGGAUAUGAGAUGAUAAGGUUACAUGCAACAUCAUUUGUUGCCUUGACAAAUGAUGUGUUAUUGUGAUA-UGUUUCAGAAGCCAAACUCAA ......-...((((((....))).)))...((...(((((((.....((((((.(((.(((((((((((.....))))))))))))))...)))))).-)))))))....))........ ( -30.70) >DroAna_CAF1 44375 114 - 1 CGCCACCAACGGACUGU--GGGUCUGCAACGGAUGUGAGAUGAUAAGGUUACAUGCAACAUCGUUUGUUUGCCUGACAAAUGAUGUGUUAUUGUGAUAGUGUUUCAAAAGCCGAAC---- .........(((((...--..)))))...(((...(((((((.....((((((.(((.(((((((((((.....))))))))))))))...))))))..)))))))....)))...---- ( -35.20) >consensus UGCCAA_ACCGUAACGUAACCGUCUGCAACGGAUAUGAGAUGAUAAGGUUACAUGCAACAUCAUUUGUUGCCUUGACAAAUGAUGUGUUAUUGUGAUA_UGUUUCAGAAGCCAAACUCGA .....................(((((...))))).(((((((.....((((((.(((.(((((((((((.....))))))))))))))...))))))..))))))).............. (-25.58 = -25.45 + -0.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:05 2006