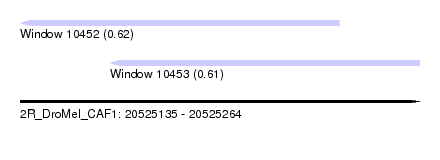
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,525,135 – 20,525,264 |
| Length | 129 |
| Max. P | 0.618190 |

| Location | 20,525,135 – 20,525,238 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.07 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -15.05 |
| Energy contribution | -14.42 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.68 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
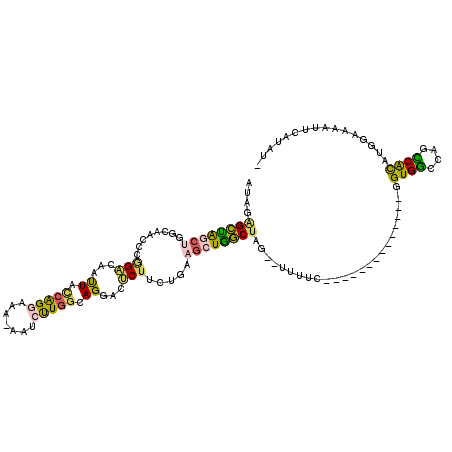
>2R_DroMel_CAF1 20525135 103 - 20766785 AUAAAGCUGGCUAGCAACUCAGACAAUUACCAGGAAA-AAUCUUGGCAGGACUCUUCUGAAGCUGGCUAG--UUUUC--------------UGUGGGCAGUCACAUGGAAAAUUCAUAUU .....((((((((((...(((((...((.((((((..-..)))))).))......))))).)))))))))--)((((--------------((((........))))))))......... ( -32.50) >DroSec_CAF1 26914 102 - 1 AUAGAGCUGGCUGGCAACUCGGACAACUACCAGAAAA-AAUCUUGGCAGGACUCUUCUGAAGCUGGCUAG--UUUUC--------------UGUGGCCAGUCACAUGGAAAGUUCAUAU- ...((((((((((((....((((.(((((((((....-........(((((...)))))...)))).)))--)).))--------------))..)))))))........)))))....- ( -31.66) >DroSim_CAF1 26848 102 - 1 AUAGAGCUGGCUGGCAACUCGGACAAUUUGCAGGAAA-AAUCUUGGCAGGACUCUUCUGAAGCUGGCUAG--UUUUC--------------UGUGACCAGUCACAUGGAAAGUUCAUUU- ...((((((((..((...(((((...(((((((((..-..)))).))))).....))))).))..)))).--(((((--------------(((((....))))).)))))))))....- ( -31.70) >DroEre_CAF1 27248 103 - 1 AUGGAGCCAACGUGCAACCCGGACAAUUACCAGGAAAGAAACUUGGCAGGACUCUUCUGAAGCUGGCUAG--UUUUC--------------GGUGGCCAGCCACAUGGAAAAUUCGUAU- ((((((((((.((....((.((.......)).))......))))))).....(((..((..((((((((.--(....--------------).))))))))..)).)))...)))))..- ( -27.00) >DroYak_CAF1 27289 102 - 1 AUAGGGCUAACUGGCAACCCUGGCAACUACCAGGAAA-AAACUUGGCAGGACCCUUCUAAAGCUGGCUAG--UUUUC--------------GGUGGCCAGCCAUAUGGAAAAUUCGUAU- ...(((....(((.(((.(((((......)))))...-....))).)))..)))(((((..((((((((.--(....--------------).))))))))....))))).........- ( -31.30) >DroAna_CAF1 26993 107 - 1 CUAGAGGAAG--UG----CCGGCCAAUUAUGGG---C-AGCCCUGGCAGGACUCUCGCAGGGCUUUCCAGGGUUUUCGUGGAGCAACAAUUGGUGGGCAGCCGCAUGGAAAAUUGGU--- (((..((...--((----((.(((((((...((---.-((((((((.(((.(((.....)))))).)))))))).))((......))))))))).)))).))...))).........--- ( -40.70) >consensus AUAGAGCUAGCUGGCAACCCGGACAAUUACCAGGAAA_AAUCUUGGCAGGACUCUUCUGAAGCUGGCUAG__UUUUC______________GGUGGCCAGCCACAUGGAAAAUUCAUAU_ ....((((((((........(((...((.(((((.......))))).))...))).....))))))))........................((((....))))................ (-15.05 = -14.42 + -0.63)

| Location | 20,525,164 – 20,525,264 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.92 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -11.03 |
| Energy contribution | -11.95 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
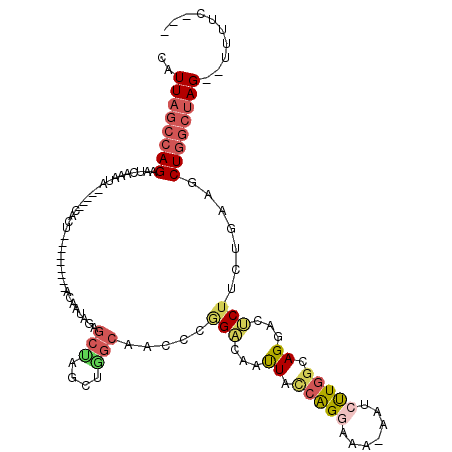
>2R_DroMel_CAF1 20525164 100 - 20766785 CAUUAGCCAGAAUAAAAUA-----CGCG---------ACCAUAAAGCUGGCUAGCAACUCAGACAAUUACCAGGAAA-AAUCUUGGCAGGACUCUUCUGAAGCUGGCUAG--UUUUC--- ...................-----....---------.....(((((((((((((...(((((...((.((((((..-..)))))).))......))))).)))))))))--)))).--- ( -28.00) >DroSec_CAF1 26942 100 - 1 CAUUAGCCAGAAUCAAAUA-----CGCA---------ACAAUAGAGCUGGCUGGCAACUCGGACAACUACCAGAAAA-AAUCUUGGCAGGACUCUUCUGAAGCUGGCUAG--UUUUC--- ...................-----....---------.....(((((((((..((...(((((...((.((((....-....)))).))......))))).))..)))))--)))).--- ( -24.30) >DroSim_CAF1 26876 100 - 1 CAUUAGCCAGAAUCAAAUA-----CGAA---------ACAAUAGAGCUGGCUGGCAACUCGGACAAUUUGCAGGAAA-AAUCUUGGCAGGACUCUUCUGAAGCUGGCUAG--UUUUC--- ...................-----....---------.....(((((((((..((...(((((...(((((((((..-..)))).))))).....))))).))..)))))--)))).--- ( -25.90) >DroEre_CAF1 27276 104 - 1 CAUUAUCCAGAGUCAAACA--GCCCACU---------ACAAUGGAGCCAACGUGCAACCCGGACAAUUACCAGGAAAGAAACUUGGCAGGACUCUUCUGAAGCUGGCUAG--UUUUC--- ..............(((((--(((..((---------.((..((((((...((.(.....).)).....(((((.......)))))..)).))))..)).))..)))).)--)))..--- ( -23.20) >DroYak_CAF1 27317 103 - 1 CAUUAGCCAGAAUCAAACA--GCCCACU---------ACAAUAGGGCUAACUGGCAACCCUGGCAACUACCAGGAAA-AAACUUGGCAGGACCCUUCUAAAGCUGGCUAG--UUUUC--- .(((((((((........(--((((...---------......)))))..(((.(((.(((((......)))))...-....))).))).............))))))))--)....--- ( -32.20) >DroAna_CAF1 27030 110 - 1 CAUUAGCCAGAAUAAUGGAAGGCCCACUUCCCACAUCCCACUAGAGGAAG--UG----CCGGCCAAUUAUGGG---C-AGCCCUGGCAGGACUCUCGCAGGGCUUUCCAGGGUUUUCGUG .....(((...(((((....((((((((((((...........).)))))--))----..)))).))))).))---)-((((((((.(((.(((.....)))))).))))))))...... ( -41.50) >consensus CAUUAGCCAGAAUCAAAUA_____CACU_________ACAAUAGAGCUAGCUGGCAACCCGGACAAUUACCAGGAAA_AAUCUUGGCAGGACUCUUCUGAAGCUGGCUAG__UUUUC___ ..((((((((...................................(((....))).....(((...((.(((((.......))))).))...))).......)))))))).......... (-11.03 = -11.95 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:58 2006