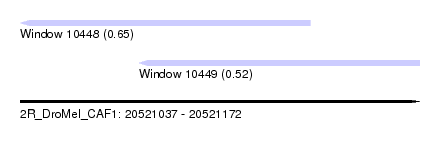
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,521,037 – 20,521,172 |
| Length | 135 |
| Max. P | 0.649360 |

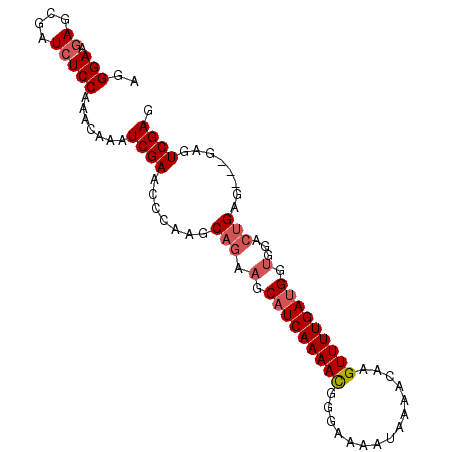
| Location | 20,521,037 – 20,521,135 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 84.83 |
| Mean single sequence MFE | -24.59 |
| Consensus MFE | -15.52 |
| Energy contribution | -15.84 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20521037 98 - 20766785 GAAGCAUCAAAAUUUGAAUAUAAAACAAGUUUUGACGGC-----GGACUGAGGAGGAGUCGAGAACUUUGCGAGUGGACCACGCAAAUGAGCAAAUCUAAUAU ...((.((((((((((.........))))))))))((((-----...((....))..)))).....((((((.(.....).))))))...))........... ( -19.40) >DroSec_CAF1 22758 94 - 1 GAAGCAUCAAAACGGGA-AAUAAAACAAGUUUUGAUGGU-----GGACUGAG---GAGUCGAGAGCUUUGCGAGUGGACCACGCGAAUGAGCAAAUCUAACAA ....(((((((((..(.-.......)..)))))))))((-----((((((..---(((.(....))))..).)))...))))((......))........... ( -22.60) >DroSim_CAF1 22702 95 - 1 GAAGCAUCAAAACGGGAAAAUAAAACAAGUUUUGAUGGU-----GGACUGAG---GAGUCGAGAGCUUUGCGAGUGGACCACGCGAAUGAGCAAAUCUAAUAA ....(((((((((..(.........)..)))))))))((-----((((((..---(((.(....))))..).)))...))))((......))........... ( -21.80) >DroEre_CAF1 22967 95 - 1 AAACCAUCAAAAUAUAUAAAUAAAACAAGUUUUGAUGGU-----GGGCCGCG---GAGUCGAGGGCUUAGCGAGUGGACCCAGCGAAUGAGCAAAUCGCAUAA ..(((((((((((...............)))))))))))-----(((((((.---((((.....)))).....)))).))).((((.(.....).)))).... ( -30.16) >DroYak_CAF1 22982 100 - 1 GAAGCAUCAAAAUGUGUAAAUAAAACAAGUUUUGAUGGUUUGGCGGGCUGUG---GAGUCGAGGGCUUUGCGAGUGGACCUCGCGAAUGAGCAAAUCGAAUAA (((.(((((((((.(((.......))).))))))))).)))...........---...((((..((((((((((.....))))))...))))...)))).... ( -29.00) >consensus GAAGCAUCAAAAUGUGAAAAUAAAACAAGUUUUGAUGGU_____GGACUGAG___GAGUCGAGAGCUUUGCGAGUGGACCACGCGAAUGAGCAAAUCUAAUAA ....(((((((((...............)))))))))........((((.......))))((..((((.....(((.....)))....))))...))...... (-15.52 = -15.84 + 0.32)

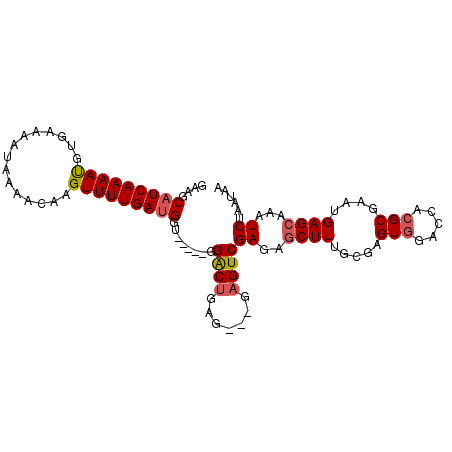
| Location | 20,521,077 – 20,521,172 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 86.99 |
| Mean single sequence MFE | -22.91 |
| Consensus MFE | -13.42 |
| Energy contribution | -14.42 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20521077 95 - 20766785 AGGGAAGAGCGAUCUCCAAACAAAUCGAACCCAAGCAGAAGCAUCAAAAUUUGAAUAUAAAACAAGUUUUGACGGCGGACUGAGGAGGAGUCGAG ..(((.((....))))).......((((..((...(((..((.((((((((((.........))))))))))..))...)))....))..)))). ( -23.40) >DroSec_CAF1 22798 91 - 1 AGGGAAGAGCGAUCUCCAAACAAAUCGAACCCAAGCAGAAGCAUCAAAACGGGA-AAUAAAACAAGUUUUGAUGGUGGACUGAG---GAGUCGAG ..(((.((....))))).......((((..((...(((.(.(((((((((..(.-.......)..))))))))).)...))).)---)..)))). ( -24.20) >DroSim_CAF1 22742 92 - 1 AGGGAAGAGCGAUCUCCAAACAAAUCGAACCCAAGCAGAAGCAUCAAAACGGGAAAAUAAAACAAGUUUUGAUGGUGGACUGAG---GAGUCGAG ..(((.((....))))).......((((..((...(((.(.(((((((((..(.........)..))))))))).)...))).)---)..)))). ( -23.40) >DroEre_CAF1 23007 92 - 1 AGGGAACAGCCAUCUCCAGACAAAUCGAAACCAAACCAAACCAUCAAAAUAUAUAAAUAAAACAAGUUUUGAUGGUGGGCCGCG---GAGUCGAG ..(((.........))).......((((..((...((..(((((((((((...............))))))))))).))....)---)..)))). ( -20.66) >consensus AGGGAAGAGCGAUCUCCAAACAAAUCGAACCCAAGCAGAAGCAUCAAAACGGGAAAAUAAAACAAGUUUUGAUGGUGGACUGAG___GAGUCGAG ..(((.((....))))).......((((.......(((.(.(((((((((...............))))))))).)...)))........)))). (-13.42 = -14.42 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:55 2006