
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,520,773 – 20,520,885 |
| Length | 112 |
| Max. P | 0.643539 |

| Location | 20,520,773 – 20,520,885 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.87 |
| Mean single sequence MFE | -40.57 |
| Consensus MFE | -28.01 |
| Energy contribution | -29.70 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20520773 112 + 20766785 AAUGGGGGCCGUGGAACUUUGCAUUUACGACUCGGC--------AGAGCUGUAAGUGCUUCCGGGCUGCCCCCCGAGGGCAAGAAUCGAUUUUCGAUGUUAAUCGAGUUGAAAACCCUGC ...(((((((.(((((....(((((((((.(((...--------.))).)))))))))))))))))(((((.....)))))....(((((..(((((....))))))))))...)))).. ( -44.60) >DroSec_CAF1 22492 111 + 1 AAUGGCGGCCGUGGAACUUCGCAUUUACGACUCGGC--------AGAGCUUUAAGUGCUUCCUGGCU-CCCCCCGAGGGCAAGAAUCGAUUUUCGAUGUUAAUCGAGUUGAAAACCCCGC ...((.(((((.((((....(((((((...(((...--------.)))...))))))))))))))))-.))..((.(((......(((((..(((((....))))))))))...))))). ( -33.50) >DroSim_CAF1 22436 111 + 1 AAUGGGGGCCGUGGAACUUCGCAUUUACGACUCGGC--------AGAGCUGUAAGUGCUUCCGGGCU-CCCCCCGAGGGCAAGAAUCGAUUUUCGAUGUUAAUCGAGUUGAAAACCCCGC ...(((((((.(((((....(((((((((.(((...--------.))).))))))))))))))))))-)))..((.(((......(((((..(((((....))))))))))...))))). ( -44.60) >DroEre_CAF1 22708 111 + 1 AAUGGGGGCCGUGGAACAUCGCAUUUACGACUCGGC--------UGGGCUGUAAGUGCUUCCGGGCU-CCCCCCGAGGGCAAGAAUCGAUUUUCGAUGUUAAUCGAGUUGAAAACCCCGC ...(((((((.(((((....(((((((((.(((...--------.))).))))))))))))))))))-)))..((.(((......(((((..(((((....))))))))))...))))). ( -42.60) >DroYak_CAF1 22725 111 + 1 AAUGGGGACCGUGGUACAUCGCAUUUACGACUCGGC--------AGAGCUGUAAGUGCUUCCGGGCU-CCCCCCGAGGGCAUGAAUCGAUUUUCGAUGUUAAUCGAGUUGAACACCCCGC ...((((...((((....)))).....((((((((.--------..(((.....(((((..((((..-...))))..)))))..((((.....)))))))..))))))))....)))).. ( -40.20) >DroAna_CAF1 21695 100 + 1 C--CGGGCUUGGGGAGUUCU---GUUGUGGCUCCGCCGGCCUCCGGAGUUGGAUGUGCUUCCGGG---------------GGGAAUCGAUUUUCGAUGUUAAUCGAGUUGAAUACCCUGC .--.(((((.(.((((((..---.....)))))).).)))))(((((((.......)).)))))(---------------(((..(((((..(((((....))))))))))...)))).. ( -37.90) >consensus AAUGGGGGCCGUGGAACUUCGCAUUUACGACUCGGC________AGAGCUGUAAGUGCUUCCGGGCU_CCCCCCGAGGGCAAGAAUCGAUUUUCGAUGUUAAUCGAGUUGAAAACCCCGC ...(((((((.(((((....((((((((..(((............)))..))))))))))))))))...(((....)))......(((((..(((((....))))))))))...)))).. (-28.01 = -29.70 + 1.69)

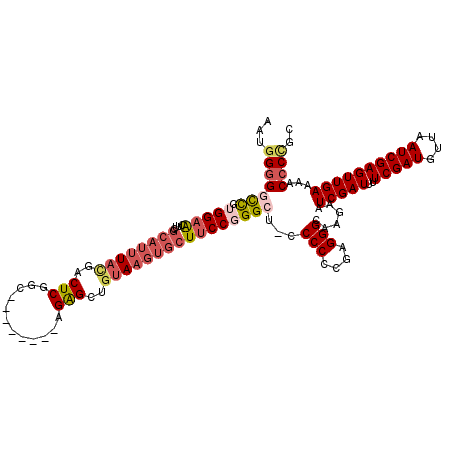
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:53 2006