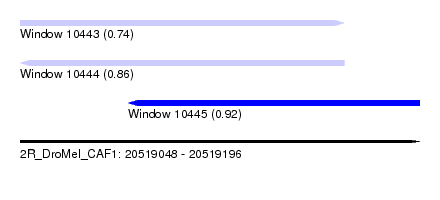
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,519,048 – 20,519,196 |
| Length | 148 |
| Max. P | 0.920842 |

| Location | 20,519,048 – 20,519,168 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.82 |
| Mean single sequence MFE | -39.42 |
| Consensus MFE | -29.42 |
| Energy contribution | -30.90 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20519048 120 + 20766785 GCCGUUUUGCCGACCGAGUACUUUUCGAGUUCGAUGGUAAUCGAUGCCUUCCGAUUCCAUUCGGACAUAUCGAUGCAUCGAUAUGUCGUAGUUCACACGACGGCCGCGGCAGCGCUUUCU (((((...(((.((((((.....)))).))((((((...(((((((...(((((......)))))..))))))).))))))...(((((.(....))))))))).))))).......... ( -43.00) >DroSec_CAF1 20714 120 + 1 GCCGUUUUGCCGACCGAGUACUUUUCGAGUCCGAUGGUAAUCGAUGCCUUCCGAUUCCGUUCGGACAUAUCGAUGCAUCGAUAUGUUUUAGUACACAAGGCGGCCACGGCAGCGCUUUGA (((((...((((.((..(((((......(((((((((.(((((........)))))))).))))))(((((((....))))))).....)))))....)))))).))))).......... ( -46.80) >DroSim_CAF1 20652 120 + 1 GCCGUUUUGCCGACCGAGUACUUUUCGAGUCCGAUGGUAAUCGAUGCCUUCCGAUUCCGUUCGGACAUAUCGAUGCAUCGAUAUGUUUUAGUACACAAGGCGGCCACGGCAGCGCUUUGA (((((...((((.((..(((((......(((((((((.(((((........)))))))).))))))(((((((....))))))).....)))))....)))))).))))).......... ( -46.80) >DroEre_CAF1 20891 110 + 1 G--------CCGACCGAGUACUUUUCGAGUUCGAUGGUAAUCGAUGCCUUCCGAUUUCUUCCACA--UAUCGAUGCAUCGAUACGAUUUAGUUCGUCACGCGGCCGCAGCAGCGCUUUGU (--------(((......((((..(((....))).))))((((((((....((((..........--.))))..))))))))((((......))))....))))(((....)))...... ( -28.30) >DroYak_CAF1 20914 119 + 1 GUCGUUUAGCCGACCGAGUACUUUUCGAGUUCGAUGGUAAUCGAUGCCUUCCGAUUACCUUCGCAC-UAUCGAUGCAUCGAUUCUAUUUAGUUCAUCACGCGGCCGCAGCAGCGCUUUGU ........((((..((((.....))))(((.(((.((((((((........)))))))).))).))-)...((((.((..(......)..)).))))...))))(((....)))...... ( -32.20) >consensus GCCGUUUUGCCGACCGAGUACUUUUCGAGUUCGAUGGUAAUCGAUGCCUUCCGAUUCCGUUCGGACAUAUCGAUGCAUCGAUAUGUUUUAGUUCACAAGGCGGCCGCGGCAGCGCUUUGU (((((...((((((((((.....)))).)))(((((...(((((((...(((((......)))))..))))))).))))).....................))).))))).......... (-29.42 = -30.90 + 1.48)

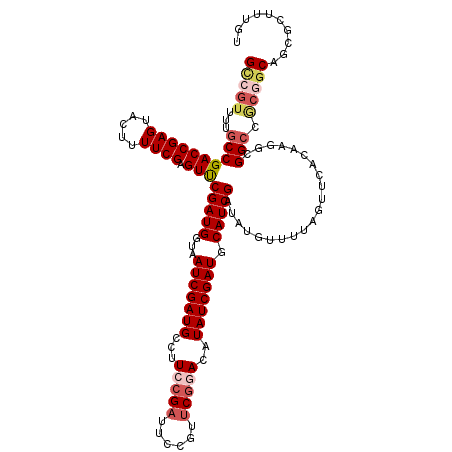

| Location | 20,519,048 – 20,519,168 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.82 |
| Mean single sequence MFE | -39.98 |
| Consensus MFE | -28.18 |
| Energy contribution | -27.94 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20519048 120 - 20766785 AGAAAGCGCUGCCGCGGCCGUCGUGUGAACUACGACAUAUCGAUGCAUCGAUAUGUCCGAAUGGAAUCGGAAGGCAUCGAUUACCAUCGAACUCGAAAAGUACUCGGUCGGCAAAACGGC ..........((((..(((((((.(((......((((((((((....))))))))))(((.(((((((((......)))))).))))))...........))).))).))))....)))) ( -41.70) >DroSec_CAF1 20714 120 - 1 UCAAAGCGCUGCCGUGGCCGCCUUGUGUACUAAAACAUAUCGAUGCAUCGAUAUGUCCGAACGGAAUCGGAAGGCAUCGAUUACCAUCGGACUCGAAAAGUACUCGGUCGGCAAAACGGC ..........(((((.((((((....(((((.....(((((((....)))))))((((((..((((((((......)))))).)).))))))......)))))..)).))))...))))) ( -44.90) >DroSim_CAF1 20652 120 - 1 UCAAAGCGCUGCCGUGGCCGCCUUGUGUACUAAAACAUAUCGAUGCAUCGAUAUGUCCGAACGGAAUCGGAAGGCAUCGAUUACCAUCGGACUCGAAAAGUACUCGGUCGGCAAAACGGC ..........(((((.((((((....(((((.....(((((((....)))))))((((((..((((((((......)))))).)).))))))......)))))..)).))))...))))) ( -44.90) >DroEre_CAF1 20891 110 - 1 ACAAAGCGCUGCUGCGGCCGCGUGACGAACUAAAUCGUAUCGAUGCAUCGAUA--UGUGGAAGAAAUCGGAAGGCAUCGAUUACCAUCGAACUCGAAAAGUACUCGGUCGG--------C ......(((....)))(((((.(..(((.((....((((((((....))))))--))....))...)))..).))(((((.(((..(((....)))...))).))))).))--------) ( -31.10) >DroYak_CAF1 20914 119 - 1 ACAAAGCGCUGCUGCGGCCGCGUGAUGAACUAAAUAGAAUCGAUGCAUCGAUA-GUGCGAAGGUAAUCGGAAGGCAUCGAUUACCAUCGAACUCGAAAAGUACUCGGUCGGCUAAACGAC ....(((((....))(((((.(((.............(((((((((.(((((.-.(((....))))))))...)))))))))....(((....)))....))).))))).)))....... ( -37.30) >consensus ACAAAGCGCUGCCGCGGCCGCCUUGUGAACUAAAACAUAUCGAUGCAUCGAUAUGUCCGAACGGAAUCGGAAGGCAUCGAUUACCAUCGAACUCGAAAAGUACUCGGUCGGCAAAACGGC .......(((((((.(..(...................((((((((.(((((...(((....))))))))...)))))))).....(((....)))...)..).))).))))........ (-28.18 = -27.94 + -0.24)

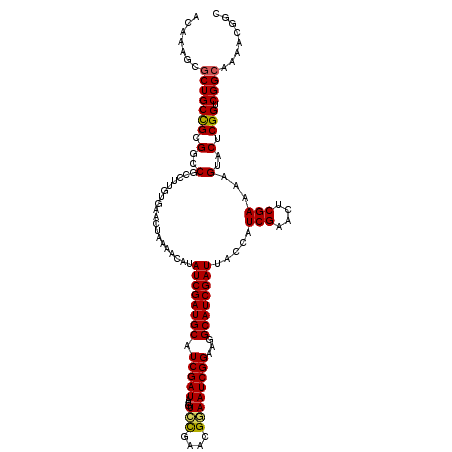
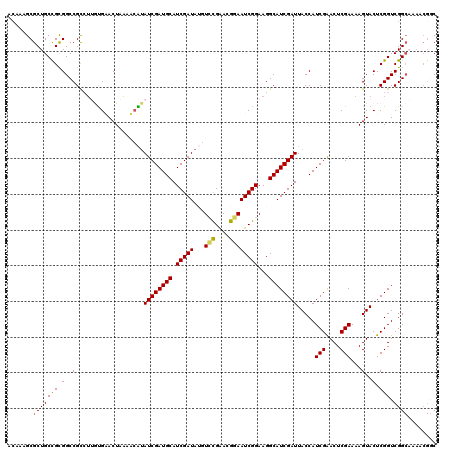
| Location | 20,519,088 – 20,519,196 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.90 |
| Mean single sequence MFE | -33.76 |
| Consensus MFE | -20.90 |
| Energy contribution | -21.58 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20519088 108 - 20766785 GUG------------UUGACAUAUCGAUGUAUGUGUUUUCAGAAAGCGCUGCCGCGGCCGUCGUGUGAACUACGACAUAUCGAUGCAUCGAUAUGUCCGAAUGGAAUCGGAAGGCAUCGA (((------------((..(((((((((((((((((((.....)))))).((....)).((((((....).)))))......)))))))))))))(((((......))))).)))))... ( -37.80) >DroSec_CAF1 20754 102 - 1 GUG----------ACUUCACAAUUCGAUGAG--------CUCAAAGCGCUGCCGUGGCCGCCUUGUGUACUAAAACAUAUCGAUGCAUCGAUAUGUCCGAACGGAAUCGGAAGGCAUCGA (((----------....)))...((((((.(--------(.(((.(((..((....))))).))).)).((....((((((((....))))))))(((((......))))))).)))))) ( -32.50) >DroSim_CAF1 20692 102 - 1 GUG----------ACUUCACAAUUCGAUGAG--------CUCAAAGCGCUGCCGUGGCCGCCUUGUGUACUAAAACAUAUCGAUGCAUCGAUAUGUCCGAACGGAAUCGGAAGGCAUCGA (((----------....)))...((((((.(--------(.(((.(((..((....))))).))).)).((....((((((((....))))))))(((((......))))))).)))))) ( -32.50) >DroEre_CAF1 20923 110 - 1 UUGGUAAAAUAUGUAUGGAUGUGUCGAUAAG--------CACAAAGCGCUGCUGCGGCCGCGUGACGAACUAAAUCGUAUCGAUGCAUCGAUA--UGUGGAAGAAAUCGGAAGGCAUCGA .........(((((((.(((((((((((...--------......((((.((....)).)))).((((......))))))))))))))).)))--))))...((..((....))..)).. ( -34.20) >DroYak_CAF1 20954 109 - 1 UUGG--AUGUGUGUCUUAACAACUCGGUAAG--------CACAAAGCGCUGCUGCGGCCGCGUGAUGAACUAAAUAGAAUCGAUGCAUCGAUA-GUGCGAAGGUAAUCGGAAGGCAUCGA ((((--...((((.((((.(.....).))))--------))))..((((.((....)).))))......))))......(((((((.(((((.-.(((....))))))))...))))))) ( -31.80) >consensus GUG__________ACUUCACAAUUCGAUGAG________CACAAAGCGCUGCCGCGGCCGCCUUGUGAACUAAAACAUAUCGAUGCAUCGAUAUGUCCGAACGGAAUCGGAAGGCAUCGA .......................(((((..................(((....)))(((................((((((((....))))))))(((((......))))).)))))))) (-20.90 = -21.58 + 0.68)

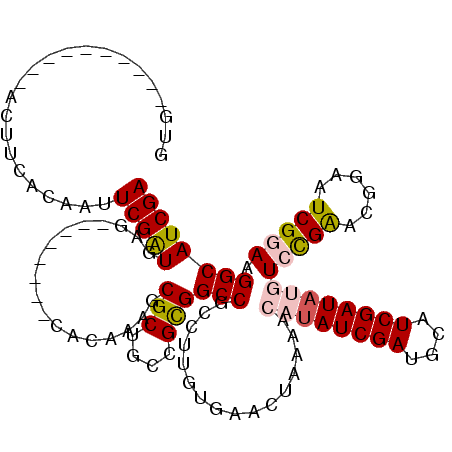
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:51 2006