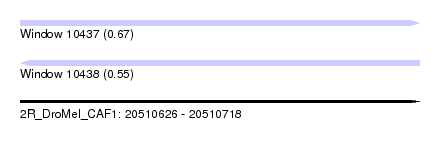
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,510,626 – 20,510,718 |
| Length | 92 |
| Max. P | 0.671253 |

| Location | 20,510,626 – 20,510,718 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 90.65 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -20.12 |
| Energy contribution | -19.68 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
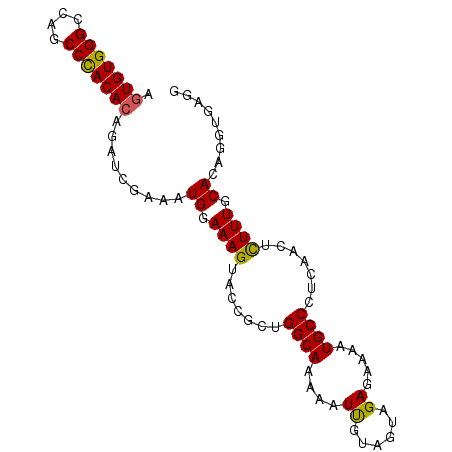
>2R_DroMel_CAF1 20510626 92 + 20766785 AGUGUGGGCCAGCCCACACAAAUCGAAAUGGAAAGUACCGCUGGCAAUUAUUGUAGUAGAGAAAAUGCCCUCAACUUUUUGCACAGGUGAGU .(((((((....)))))))................((((....((((.......(((.(((........))).)))..))))...))))... ( -25.50) >DroSec_CAF1 12097 92 + 1 AGUGUGGGCCAGCCCACACAGAUCGAAAUGGAAAGUACCGCUGGCAAAAAUUGUAGUAGAGAAAAUGCCCUCAACUCUUUGCAUAGGUAAGG .(((((((....)))))))................((((....(((((....(.(((.(((........))).)))))))))...))))... ( -26.10) >DroSim_CAF1 12091 92 + 1 AGUGUGGGCCAGCCCACACAGAUCGAAAUGGAAAGUACCGCUGGCAAAAAUUGUAGUAGAGAAAAUGCCCUCAACUCUUUGCACAGGUGAGG .(((((((....)))))))................((((....(((((....(.(((.(((........))).)))))))))...))))... ( -25.80) >DroEre_CAF1 12057 92 + 1 AGUGUGGGCCAGCCUACACAGAUCGAAAUGGAAAGUACUGCUGGCAGAAAUCAUAGUAGAGAAAAUGCCUUCAACACUUUGCACGGCUGAGG .(((((((....)))))))((.(((...((.(((((..((..((((....((......)).....))))..))..))))).))))))).... ( -26.70) >DroYak_CAF1 12049 92 + 1 AGUGUGGGGCAGCCUACAGAGAUCGAAAUGGAAAGUACCGCAGGCAAAAAUCAUAGUAGAGAAAAUGCCUUCAACACUUUGCACAGGUGAGG ..((((((....))))))....(((...((.(((((...(.(((((....((......)).....))))).)...))))).))....))).. ( -20.00) >consensus AGUGUGGGCCAGCCCACACAGAUCGAAAUGGAAAGUACCGCUGGCAAAAAUUGUAGUAGAGAAAAUGCCCUCAACUCUUUGCACAGGUGAGG .(((((((....))))))).........((.((((.......((((....((......)).....)))).......)))).))......... (-20.12 = -19.68 + -0.44)

| Location | 20,510,626 – 20,510,718 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 90.65 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -17.87 |
| Energy contribution | -18.35 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
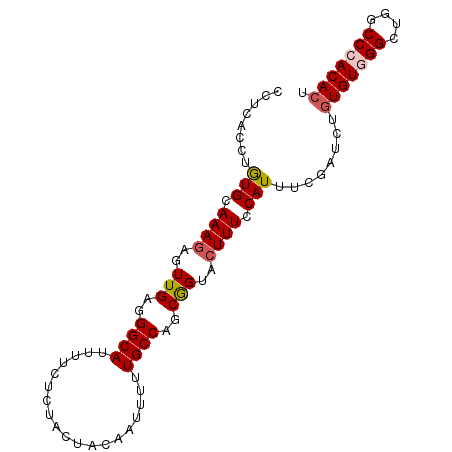
>2R_DroMel_CAF1 20510626 92 - 20766785 ACUCACCUGUGCAAAAAGUUGAGGGCAUUUUCUCUACUACAAUAAUUGCCAGCGGUACUUUCCAUUUCGAUUUGUGUGGGCUGGCCCACACU .((((.((........)).))))((((...................))))...((......))..........(((((((....))))))). ( -24.21) >DroSec_CAF1 12097 92 - 1 CCUUACCUAUGCAAAGAGUUGAGGGCAUUUUCUCUACUACAAUUUUUGCCAGCGGUACUUUCCAUUUCGAUCUGUGUGGGCUGGCCCACACU ...((((...(((((((((.(((((....))))).))).....))))))....))))................(((((((....))))))). ( -24.70) >DroSim_CAF1 12091 92 - 1 CCUCACCUGUGCAAAGAGUUGAGGGCAUUUUCUCUACUACAAUUUUUGCCAGCGGUACUUUCCAUUUCGAUCUGUGUGGGCUGGCCCACACU ......(((.(((((((((.(((((....))))).))).....))))))))).((......))..........(((((((....))))))). ( -26.20) >DroEre_CAF1 12057 92 - 1 CCUCAGCCGUGCAAAGUGUUGAAGGCAUUUUCUCUACUAUGAUUUCUGCCAGCAGUACUUUCCAUUUCGAUCUGUGUAGGCUGGCCCACACU ..((((((.((((((((((((..((((...((........))....))))..))))))))((......))....))))))))))........ ( -21.80) >DroYak_CAF1 12049 92 - 1 CCUCACCUGUGCAAAGUGUUGAAGGCAUUUUCUCUACUAUGAUUUUUGCCUGCGGUACUUUCCAUUUCGAUCUCUGUAGGCUGCCCCACACU ..............(((((....((((.....((......)).....((((((((.....((......))...))))))))))))..))))) ( -20.60) >consensus CCUCACCUGUGCAAAGAGUUGAGGGCAUUUUCUCUACUACAAUUUUUGCCAGCGGUACUUUCCAUUUCGAUCUGUGUGGGCUGGCCCACACU ........(((.((((..(((..((((...................))))..)))..)))).)))........(((((((....))))))). (-17.87 = -18.35 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:44 2006