
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,485,786 – 20,485,894 |
| Length | 108 |
| Max. P | 0.701673 |

| Location | 20,485,786 – 20,485,894 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.42 |
| Mean single sequence MFE | -32.08 |
| Consensus MFE | -21.77 |
| Energy contribution | -21.58 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20485786 108 - 20766785 -UUAACCCUGAAUUU----C-UU-GGUUCAGAUUCUUGCGUCUAGCCAUGGCCAGUAUCAUGGACAUACUGAAGAUCAAGCCAUUCGUUCAGGUGUCCGUGGGUCAGUUGCUUUG -......(((((((.----.-..-)))))))............(((..(((((.....(((((((((.(((((((.........)).)))))))))))))))))))...)))... ( -31.70) >DroGri_CAF1 12540 112 - 1 -UGAAUCUUGAACUUUAUUC-UU-CGGAUAGAUUUCUGCGCUUGGCCAUGGCCAGCAUUAUGGAUAUCUUGAAGAUUAAGCCGUUCGUGCAAGUGUCCGUCGGACAGCUGCUUUG -........((((((((.((-((-((...((((.((((.(((.(((....))))))....)))).)))))))))).))))..))))..(((..(((((...)))))..))).... ( -31.70) >DroEre_CAF1 9927 107 - 1 --UGACCGUGCACUU----C-UU-GGUUCAGAUUCCUGCGCCUGGCCAUGGCCAGCAUCAUGGACAUUCUGAAGAUCAAGCCAUUCGUUCAGGUGUCCGUGGGUCAGCUGCUUUG --(((((((((....----.-..-(((.(((....))).)))((((....))))))))(((((((((.(((((((.........)).)))))))))))))))))))......... ( -42.30) >DroMoj_CAF1 13103 104 - 1 -AACAUUUUGAUCUAUUU----------CAGAUUCCUGCGCUUGGCCAUGGCCAGCAUCAUGGAUAUAUUGAAGAUUAAGCCAUUCGUGCAAGUCUCUGUGGGUCAGCUGCUCUG -......(((((((((..----------.(((((..((((((((((....)))))....((((....(((...)))....))))..))))))))))..)))))))))........ ( -26.90) >DroAna_CAF1 9808 110 - 1 CUUGACCUACAACUC----CGAU-CUUUCAGAUUCCUUCGCUUGGCUAUGGCCAGCAUCAUGGACAUCCUCAAGAUCAAGCCCUUCGUUCAGGUCUCCGUCGGCCAGCUACUCUG .(((.((.((.....----.(((-(((...((((((...(((.(((....)))))).....))).)))...)))))).((.(((......))).))..)).)).)))........ ( -24.50) >DroPer_CAF1 13557 109 - 1 -UUAACCCUGAUCUU----G-CCCUUCCCAGGUUCCUGCGUCUGGCCAUGGCCAGCAUCAUGGACAUACUGAAGAUCAAGCCGUUCGUUCAGGUGUCCGUGGGACAGCUGCUCUG -..............----.-.......(((((..(((((.(((((....)))))).((((((((((.(((((((.(.....).)).)))))))))))))))).)))..)).))) ( -35.40) >consensus _UUAACCUUGAACUU____C_UU_CGUUCAGAUUCCUGCGCCUGGCCAUGGCCAGCAUCAUGGACAUACUGAAGAUCAAGCCAUUCGUUCAGGUGUCCGUGGGUCAGCUGCUCUG ............................((((.....(((.(((((....)))))).((((((((((.(((((((.........)).)))))))))))))))....))...)))) (-21.77 = -21.58 + -0.19)

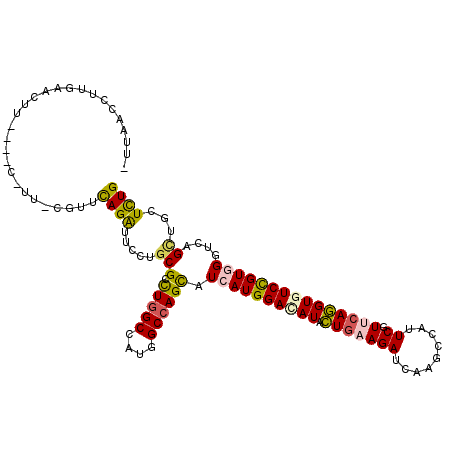
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:37 2006