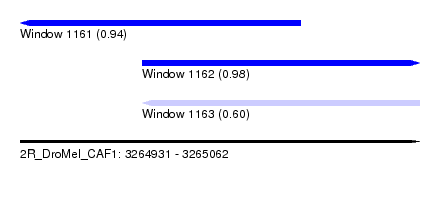
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,264,931 – 3,265,062 |
| Length | 131 |
| Max. P | 0.984985 |

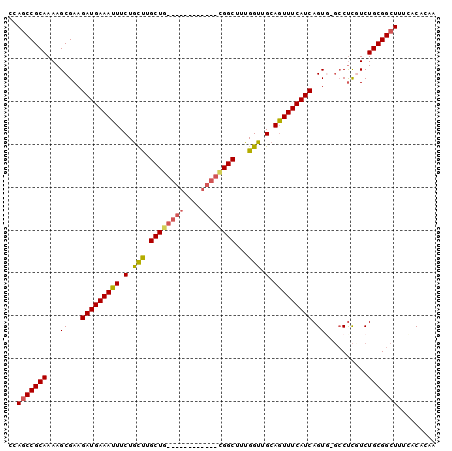
| Location | 3,264,931 – 3,265,023 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 85.33 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -22.82 |
| Energy contribution | -23.66 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937155 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3264931 92 - 20766785 CCAGCCGCAAAAGCGAAGAUGAAAUUUCUGCUUGCU--------UGCUGCGGCUUUGGUUGCAGUUUCAUCAGUGUGCCUCCUCUGCGGCUUUCACACAA ..(((((((...(((..((((((((....((.....--------.))((((((....))))))))))))))..)))........)))))))......... ( -28.50) >DroSec_CAF1 34022 86 - 1 UCAGCCGCAAAAGCGAAGAUGAAAUUUCUGCCUGCUG------------CGGCUUUGGUUGCAAUUUCAUCAGU--GCCUCGUCUGCGGCUUUCACACAA ..(((((((.(.(((..(((((((((.(.(((.((..------------..))...))).).)))))))))..)--))....).)))))))......... ( -29.70) >DroSim_CAF1 43393 98 - 1 CCAGCCGCAAAAGCGAAGAUGAAAUUUCUACCUGCUGCGGCUGCGGCUGCGGCUUUGGUUGCAGUUUCAUCAGU--GCCUCGUCUGCGGCUUUCACACAA ..(((((((.(.(((..(((((((((.(.(((.((((((((....))))))))...))).).)))))))))..)--))....).)))))))......... ( -43.20) >DroEre_CAF1 33228 88 - 1 CCAGCCGCAAAAGCGAAGAUGAAAUUUCUGUUUGCUG------------UGGCUUUGGUUGCAGUUUCAUCAGUGCGCCUCCUCUGCGGCUUUCACACAA ..(((((((...(((..((((((..........((((------------..((....))..))))))))))....)))......)))))))......... ( -28.20) >DroYak_CAF1 33300 88 - 1 CCAUCCGCAAAAGCGAAGAUGAAAUUUCUGUUUGCUG------------UGGCUUUGGUUGCAGUUUCAUCAGUGACGCCUGUCUGCGGCUUUUACACAA ......(.((((((...((((((..........((((------------..((....))..)))))))))).....(((......))))))))).).... ( -23.40) >consensus CCAGCCGCAAAAGCGAAGAUGAAAUUUCUGCUUGCUG____________CGGCUUUGGUUGCAGUUUCAUCAGUG_GCCUCGUCUGCGGCUUUCACACAA ..(((((((...((...(((((((((.(.(((.((((((((....))))))))...))).).))))))))).))..........)))))))......... (-22.82 = -23.66 + 0.84)


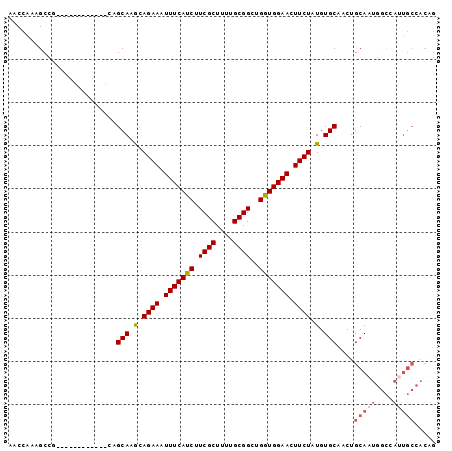
| Location | 3,264,971 – 3,265,062 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 81.23 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -19.01 |
| Energy contribution | -19.86 |
| Covariance contribution | 0.85 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3264971 91 + 20766785 AACCAAAGCCGCAGCA--------AGCAAGCAGAAAUUUCAUCUUCGCUUUUGCGGCUGGUGGAACUUCUAUGUGCAACUGCAAUGGCCAUUGCCACAG ..(((.((((((((.(--------((((((..((....))..))).))))))))))))..)))........(((((((..((....))..))).)))). ( -29.90) >DroSec_CAF1 34060 81 + 1 AACCAAAGCCG------------CAGCAGGCAGAAAUUUCAUCUUCGCUUUUGCGGCUGAUGGAACUUCUAUGUGCAACUGCAAG------UGCAACUG ..(((.(((((------------(((.((((.(((........)))))))))))))))..)))........((..(........)------..)).... ( -25.70) >DroSim_CAF1 43431 84 + 1 AACCAAAGCCGCAGCCGCAGCCGCAGCAGGUAGAAAUUUCAUCUUCGCUUUUGCGGCUGGUGGAACUUCUAUGUGCAACUGCAA--------------- ..........((((..((....)).(((.((((((.(((((((.((((....))))..))))))).)))))).)))..))))..--------------- ( -31.70) >DroEre_CAF1 33268 87 + 1 AACCAAAGCCA------------CAGCAAACAGAAAUUUCAUCUUCGCUUUUGCGGCUGGUGGAACUUCUAUGUGCAACUGCAAUGGCCAUUGCCACAG .......((..------------..(((...((((.(((((((.((((....))))..))))))).))))...)))....))..((((....))))... ( -24.50) >DroYak_CAF1 33340 87 + 1 AACCAAAGCCA------------CAGCAAACAGAAAUUUCAUCUUCGCUUUUGCGGAUGGUGGAACUUCUAUGUGCAACUGCAAUGGCCAUUGCCACAG .......((..------------..(((...((((.((((((((((((....))))).))))))).))))...)))....))..((((....))))... ( -26.20) >consensus AACCAAAGCCG____________CAGCAAGCAGAAAUUUCAUCUUCGCUUUUGCGGCUGGUGGAACUUCUAUGUGCAACUGCAAUGGCCAUUGCCACAG .........................(((.(.((((.(((((((.((((....))))..))))))).)))).).)))....(((((....)))))..... (-19.01 = -19.86 + 0.85)


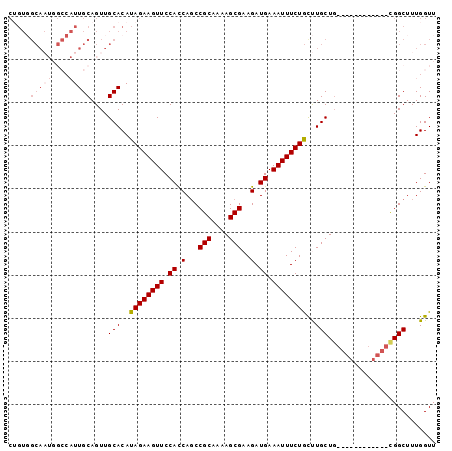
| Location | 3,264,971 – 3,265,062 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 81.23 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -14.13 |
| Energy contribution | -15.22 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3264971 91 - 20766785 CUGUGGCAAUGGCCAUUGCAGUUGCACAUAGAAGUUCCACCAGCCGCAAAAGCGAAGAUGAAAUUUCUGCUUGCU--------UGCUGCGGCUUUGGUU ((((((((((.((....)).))))).))))).....(((..(((((((.(((((((((.......)))..)))))--------)..))))))).))).. ( -34.50) >DroSec_CAF1 34060 81 - 1 CAGUUGCA------CUUGCAGUUGCACAUAGAAGUUCCAUCAGCCGCAAAAGCGAAGAUGAAAUUUCUGCCUGCUG------------CGGCUUUGGUU .(((((((------...((((..(((....((((((.((((...(((....)))..)))).)))))))))))))))------------)))))...... ( -24.50) >DroSim_CAF1 43431 84 - 1 ---------------UUGCAGUUGCACAUAGAAGUUCCACCAGCCGCAAAAGCGAAGAUGAAAUUUCUACCUGCUGCGGCUGCGGCUGCGGCUUUGGUU ---------------.(((((((((((......))......(((((((..((.(.(((.......))).)))..))))))))))))))))......... ( -28.40) >DroEre_CAF1 33268 87 - 1 CUGUGGCAAUGGCCAUUGCAGUUGCACAUAGAAGUUCCACCAGCCGCAAAAGCGAAGAUGAAAUUUCUGUUUGCUG------------UGGCUUUGGUU ((((((((((.((....)).))))).))))).....(((..(((((((...(((((.(.((....))).)))))))------------))))).))).. ( -28.80) >DroYak_CAF1 33340 87 - 1 CUGUGGCAAUGGCCAUUGCAGUUGCACAUAGAAGUUCCACCAUCCGCAAAAGCGAAGAUGAAAUUUCUGUUUGCUG------------UGGCUUUGGUU ..........(((((..((....(((.(((((((((....(((((((....)))..)))).))))))))).)))..------------..))..))))) ( -25.90) >consensus CUGUGGCAAUGGCCAUUGCAGUUGCACAUAGAAGUUCCACCAGCCGCAAAAGCGAAGAUGAAAUUUCUGCUUGCUG____________CGGCUUUGGUU .....(((((....)))))....(((..((((((((.((.(...(((....)))..).)).))))))))..)))......................... (-14.13 = -15.22 + 1.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:50 2006