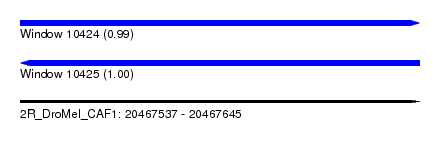
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,467,537 – 20,467,645 |
| Length | 108 |
| Max. P | 0.996111 |

| Location | 20,467,537 – 20,467,645 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.52 |
| Mean single sequence MFE | -53.02 |
| Consensus MFE | -37.61 |
| Energy contribution | -37.59 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.42 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
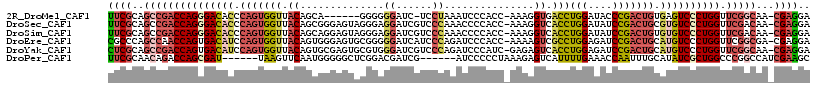
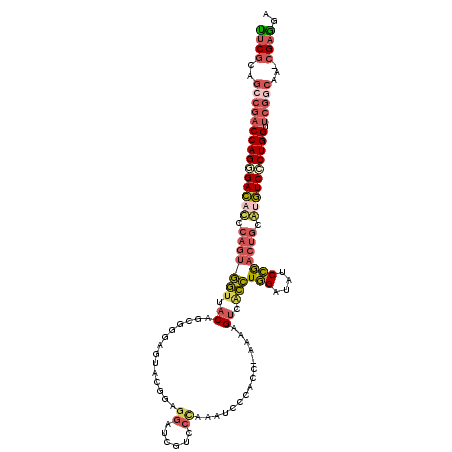
>2R_DroMel_CAF1 20467537 108 + 20766785 UCCUCG-UUGCCGAACCAGGGACUCACAGUCGGGUAUCCAGGUCACCUUU-GGUGGGAUUUAGGA-GAUCCCCCC------UGCUGUAACCACUGGGUGUCCCUGGUCGGCUGCGAA ...(((-(.(((((.((((((((..(((((.(((...(((((.....)))-)).((((((.....-)))))))))------.))))).(((....)))))))))))))))).)))). ( -53.90) >DroSec_CAF1 10243 115 + 1 UCCUCG-UUGUCGAACCAGGGACACGCAGUCGGAUAUCCAGGUGACCUUU-GGUGGGGUUUGGGACGAUCCUCCCUACUCCCGCUGUAACCACUGGGUGUCCCUGGUCGGCUGCGAA ...(((-(.(((((.((((((((((.((((.((....)).(((.((....-((((((((..((((......)))).)).)))))))).))))))).))))))))))))))).)))). ( -61.90) >DroSim_CAF1 10307 115 + 1 UCCUCG-UUGUCGAACCAGGGACACACAGUCGGAUAUCCAGGUGACCUUU-GGUGGGGUUUGGGACGAUCCUCCCUACUCCUGCUGUAACCACUGGGUGUCCCUGGUCGGCUGCGAA ...(((-(.(((((.((((((((((.((((.((.(((.((((........-(((((((...(((.....))))))))))))))..))).)))))).))))))))))))))).)))). ( -57.50) >DroEre_CAF1 10444 115 + 1 UCCUCG-UCGCCGAACCAGGGACAUGCAGUCGGAUCUCCAGGCGACUUUU-GGUGGGAUCUGGGAUGAUCCCCCGCACUCCCACUGUAACCACUGGAUGUCACUGGUUGGCUGGGCG ....((-(((((.((((((.(((((.((((.((....)).((..((....-(((((((..((((.......))))...)))))))))..)))))).))))).)))))))))..)))) ( -49.00) >DroYak_CAF1 10495 115 + 1 UCCUCG-UUGCCGAACCAGGGACAUGCAGUCGGAUCUCCAGGUGACUCUC-GAUGGGAUCUGGGACGAUCCCACGCACUCGCACUGUAACCACUGGAUGUCACUGGUCGGCUGCGAG ..((((-(.(((((.((((.(((((.((((.((.....(((((((....(-(.(((((((......)))))))))...)))).)))...)))))).))))).))))))))).))))) ( -57.10) >DroPer_CAF1 12556 105 + 1 GCUUCGAUGGCCGGGCCAGCGAUAUGCAAAUUGGUUUCAAAAUGACUCUUUAGGGGGAU------CGAUCGUCCGAGCCCCCAUUGAACUUA------AUCGCUGGUCUGUUGCGAA ...(((.....((((((((((((.....((..((((.......))))..)).(((((.(------((......))).)))))..........------))))))))))))...))). ( -38.70) >consensus UCCUCG_UUGCCGAACCAGGGACACGCAGUCGGAUAUCCAGGUGACCUUU_GGUGGGAUUUGGGACGAUCCCCCCCACUCCCACUGUAACCACUGGGUGUCCCUGGUCGGCUGCGAA ...(((...(((((.((((((((((.((((.((....)).((((.......((.((((((......)))))))).......))))......)))).)))))))))))))))..))). (-37.61 = -37.59 + -0.02)


| Location | 20,467,537 – 20,467,645 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.52 |
| Mean single sequence MFE | -46.90 |
| Consensus MFE | -27.42 |
| Energy contribution | -28.60 |
| Covariance contribution | 1.18 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20467537 108 - 20766785 UUCGCAGCCGACCAGGGACACCCAGUGGUUACAGCA------GGGGGGAUC-UCCUAAAUCCCACC-AAAGGUGACCUGGAUACCCGACUGUGAGUCCCUGGUUCGGCAA-CGAGGA ((((..(((((((((((((.(.((((((((((....------.(((((((.-......))))).))-....))))))(((....))))))).).)))))))).)))))..-)))).. ( -53.40) >DroSec_CAF1 10243 115 - 1 UUCGCAGCCGACCAGGGACACCCAGUGGUUACAGCGGGAGUAGGGAGGAUCGUCCCAAACCCCACC-AAAGGUCACCUGGAUAUCCGACUGCGUGUCCCUGGUUCGACAA-CGAGGA ((((..(.(((((((((((((.(((((((.((.(.(((.((.((((......))))..))))).).-....)).)))(((....))))))).)))))))))).))).)..-)))).. ( -47.50) >DroSim_CAF1 10307 115 - 1 UUCGCAGCCGACCAGGGACACCCAGUGGUUACAGCAGGAGUAGGGAGGAUCGUCCCAAACCCCACC-AAAGGUCACCUGGAUAUCCGACUGUGUGUCCCUGGUUCGACAA-CGAGGA ((((..(.(((((((((((((.((((((......((((....((((......))))..(((.....-...)))..)))).....)).)))).)))))))))).))).)..-)))).. ( -46.50) >DroEre_CAF1 10444 115 - 1 CGCCCAGCCAACCAGUGACAUCCAGUGGUUACAGUGGGAGUGCGGGGGAUCAUCCCAGAUCCCACC-AAAAGUCGCCUGGAGAUCCGACUGCAUGUCCCUGGUUCGGCGA-CGAGGA ..(((.(((((((((.(((((.((((((...(((.(.((.(..((((((((......)))))).))-...).)).)))).....)).)))).))))).)))))).)))..-.).)). ( -48.50) >DroYak_CAF1 10495 115 - 1 CUCGCAGCCGACCAGUGACAUCCAGUGGUUACAGUGCGAGUGCGUGGGAUCGUCCCAGAUCCCAUC-GAGAGUCACCUGGAGAUCCGACUGCAUGUCCCUGGUUCGGCAA-CGAGGA ((((..(((((((((.(((((.((((((...(((.(.((.(.(((((((((......))))))).)-)..).)).)))).....)).)))).))))).)))).)))))..-)))).. ( -54.70) >DroPer_CAF1 12556 105 - 1 UUCGCAACAGACCAGCGAU------UAAGUUCAAUGGGGGCUCGGACGAUCG------AUCCCCCUAAAGAGUCAUUUUGAAACCAAUUUGCAUAUCGCUGGCCCGGCCAUCGAAGC ((((...(.(.((((((((------..........(((((.(((......))------).)))))......(.((..(((....)))..)))..)))))))).).).....)))).. ( -30.80) >consensus UUCGCAGCCGACCAGGGACACCCAGUGGUUACAGCGGGAGUACGGAGGAUCGUCCCAAAUCCCACC_AAAAGUCACCUGGAUAUCCGACUGCAUGUCCCUGGUUCGGCAA_CGAGGA ((((..(((((((((((((((.(((((((.((..............((......))...............)).)))(((....))))))).)))))))))).)))))...)))).. (-27.42 = -28.60 + 1.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:31 2006