
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,458,189 – 20,458,300 |
| Length | 111 |
| Max. P | 0.861607 |

| Location | 20,458,189 – 20,458,300 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.35 |
| Mean single sequence MFE | -30.43 |
| Consensus MFE | -18.79 |
| Energy contribution | -19.10 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
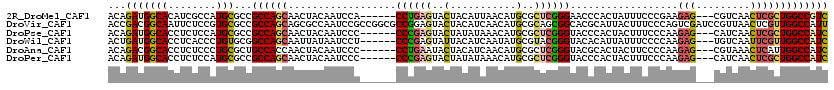
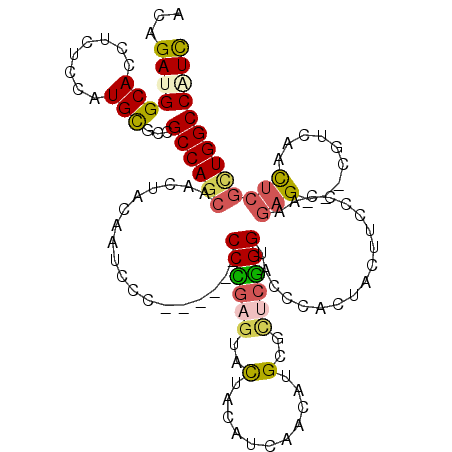
>2R_DroMel_CAF1 20458189 111 + 20766785 ACAGAUGGCACAUCGCCAUGCGCCGCCAGCAACUACAAUCCA------CCUGAGUACUACAUUAACAUGCGCUCGGGAACCCACUAUUUCCCGAAGAG---CGUCAACUCGCUGGCCGUC ....(((((.....)))))..((.((((((.(((.((.....------..))))).............(((((((((((........)))))...)))---)))......)))))).)). ( -34.10) >DroVir_CAF1 1000 120 + 1 ACCGACGGCAAUUCUCCGUGCGCCGCCAGCAGCGCCAAUCCGCCGGCGCCGGAGUACUACAUCAACAUGCGCAGCGGCACGCAUUACUUUCCCAGUCGAUCCGUUAACUCGUUGGCCAUU .((((((((....(((((.((((((......(((......)))))))))))))).....(((....))).))(((((..((.(((........)))))..)))))....))))))..... ( -36.50) >DroPse_CAF1 1006 111 + 1 ACAGAUGGCACCUCUCCAUGCGCCGCCAGCAACUACAAUCCC------CCCGAGUACUAUAUAAACAUGCGCUCGGGUACCCACUACUUUCCCAAGAG---CAUCAACUCGCUGGCCAUC ...(((((((........)))...((((((............------((((((..(...........)..))))))..................(((---......))))))))))))) ( -31.10) >DroWil_CAF1 1045 111 + 1 ACUGAUGGCACCUCACCCUGUGCGGCCAGCAAUUAUAAUCCU------CCCGAGUAUUACAUCAAUAUGCGUACGGGUACACAUUAUUUCCCCAAGAG---UGUCAAUUCGUUGGCCAUC .......((((........))))(((((((....(((((...------((((.((((.........))))...)))).....)))))........(((---(....)))))))))))... ( -28.60) >DroAna_CAF1 1032 111 + 1 ACAGACGGCACCUCUCCCUGCGCUGCCACCAACUACAAUCCC------CCUGAAUACUACAUCAACAUGCGCUCGGGUACGCACUACUUCCCCAAGAG---CGUAAACUCAUUGGCCAUC ......((((.(.(.....).).)))).((((..........------..(((........)))...((((((((((............)))...)))---))))......))))..... ( -21.20) >DroPer_CAF1 1678 111 + 1 ACAGAUGGCACCUCUCCAUGCGCCGCCAGCAACUACAAUCCC------CCCGAGUACUAUAUAAACAUGCGCUCGGGUACCCACUACUUUCCCAAGAG---CAUCAACUCGCUGGCCAUC ...(((((((........)))...((((((............------((((((..(...........)..))))))..................(((---......))))))))))))) ( -31.10) >consensus ACAGAUGGCACCUCUCCAUGCGCCGCCAGCAACUACAAUCCC______CCCGAGUACUACAUCAACAUGCGCUCGGGUACCCACUACUUCCCCAAGAG___CGUCAACUCGCUGGCCAUC ...(((((((........)))...((((((..................((((((..(...........)..))))))..................(((.........))))))))))))) (-18.79 = -19.10 + 0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:29 2006