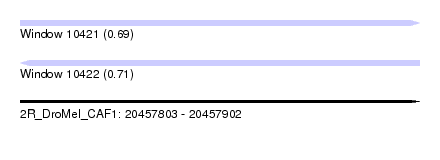
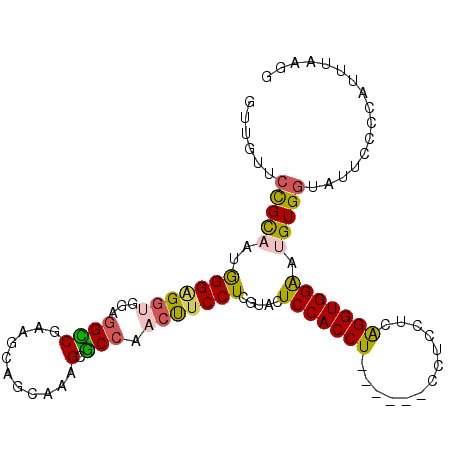
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,457,803 – 20,457,902 |
| Length | 99 |
| Max. P | 0.710145 |

| Location | 20,457,803 – 20,457,902 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.62 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -21.20 |
| Energy contribution | -21.09 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20457803 99 + 20766785 CCUUAAAUGGGGAAUACCACAUCCCACCUGAGGAGG------AGGUGGAGUACGAGGAAGUUGGCGCUUUGCUGCUUCGGCAUGCACCACCCAUUGCAGAACAAC .((..((((((.....((.(..(((((((.......------)))))).)...).))....(((.((..(((((...))))).)).)))))))))..))...... ( -29.60) >DroPse_CAF1 605 105 + 1 AGCCCAAUGGGGAGUACCACAUUCCACCCGAAGAGGAAGAUGAGGUGGAGUACGAGGAGGUGGGAGCGCUACUGUUGCGCCCCCAGCCUCCUAUAACGGACUACC ..(((....)))(((.((..((((((((((..........)).))))))))...((((((((((.((((.......)))).)))).)))))).....)))))... ( -43.80) >DroSim_CAF1 579 99 + 1 CCUUAAAUGGGGAAUACCACAUCCCACCUGAGGAGG------AGGUGGAGUAUGAGGAAGUUGGCGCUUUGCUCCUUCGGCCUGCACCCCCCAUUGCAGAACAAC .((..(((((((....((.((((((((((.......------)))))).).))).)).....((.((...(((.....)))..)).)))))))))..))...... ( -31.50) >DroEre_CAF1 579 99 + 1 CCUUAAAUGGGGAAUACCACAUUCCACCUGAGGAGG------AGGUGGAGUACGAGGAAGUUGGCGCUUUGCUGCUUCGGCCUCCAGCUCCCAUUGCGGAACAAC (((((...(((((((.....))))).))))))).((------((.(((((..(((((.(((.........))).)))))..))))).)))).............. ( -36.20) >DroYak_CAF1 579 99 + 1 CCUUAAAUGGGGAAUAUCACAUUCCACCUGAGGAGG------AGGUGGAGUAUGAGGAAGUUGGCGCUUUGCUACUUCGACCACCACCCCCCAUUGCGGAACAAC ((...(((((((....(((.(((((((((.......------))))))))).))).(((((.(((.....))))))))..........)))))))..))...... ( -36.40) >DroPer_CAF1 1277 105 + 1 AGCCCAAUGGGGAGUACCACAUUCCACCCGAAGAGGAAGAUGAGGUGGAGUACGAGGAGGUGGGAGCGCUACUGUUGCGCCCCCAGCCUCCUAUAACGGACUACC ..(((....)))(((.((..((((((((((..........)).))))))))...((((((((((.((((.......)))).)))).)))))).....)))))... ( -43.80) >consensus CCUUAAAUGGGGAAUACCACAUUCCACCUGAGGAGG______AGGUGGAGUACGAGGAAGUUGGCGCUUUGCUGCUUCGGCCUCCACCUCCCAUUGCGGAACAAC .....(((((((........((((((((...............)))))))).(((((.(((.........))).))))).........))))))).......... (-21.20 = -21.09 + -0.11)

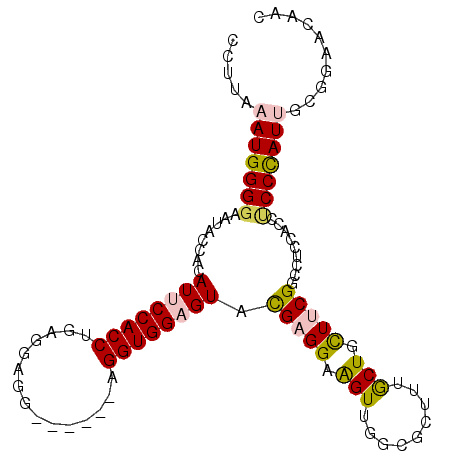
| Location | 20,457,803 – 20,457,902 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 78.62 |
| Mean single sequence MFE | -38.17 |
| Consensus MFE | -19.46 |
| Energy contribution | -18.77 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20457803 99 - 20766785 GUUGUUCUGCAAUGGGUGGUGCAUGCCGAAGCAGCAAAGCGCCAACUUCCUCGUACUCCACCU------CCUCCUCAGGUGGGAUGUGGUAUUCCCCAUUUAAGG ......((..((((((((((((.(((.......)))..))))))....((.(((.(.((((((------.......)))))))))).)).....))))))..)). ( -30.80) >DroPse_CAF1 605 105 - 1 GGUAGUCCGUUAUAGGAGGCUGGGGGCGCAACAGUAGCGCUCCCACCUCCUCGUACUCCACCUCAUCUUCCUCUUCGGGUGGAAUGUGGUACUCCCCAUUGGGCU ...(((((((...((((((.(((((((((.......)))))))))))))))...))(((((((.............)))))))..((((......)))).))))) ( -46.82) >DroSim_CAF1 579 99 - 1 GUUGUUCUGCAAUGGGGGGUGCAGGCCGAAGGAGCAAAGCGCCAACUUCCUCAUACUCCACCU------CCUCCUCAGGUGGGAUGUGGUAUUCCCCAUUUAAGG ......((..((((((((((((..(((....).))...))))).......(((((..((((((------.......))))))..)))))....)))))))..)). ( -38.40) >DroEre_CAF1 579 99 - 1 GUUGUUCCGCAAUGGGAGCUGGAGGCCGAAGCAGCAAAGCGCCAACUUCCUCGUACUCCACCU------CCUCCUCAGGUGGAAUGUGGUAUUCCCCAUUUAAGG ...(((((((...(((((.(((((..(((((..((.....))...))...)))..))))).))------)))......)))))))((((......))))...... ( -30.20) >DroYak_CAF1 579 99 - 1 GUUGUUCCGCAAUGGGGGGUGGUGGUCGAAGUAGCAAAGCGCCAACUUCCUCAUACUCCACCU------CCUCCUCAGGUGGAAUGUGAUAUUCCCCAUUUAAGG ......((..(((((((((((......(((((.((.....))..))))).(((((.(((((((------.......))))))).))))))))))))))))...)) ( -36.00) >DroPer_CAF1 1277 105 - 1 GGUAGUCCGUUAUAGGAGGCUGGGGGCGCAACAGUAGCGCUCCCACCUCCUCGUACUCCACCUCAUCUUCCUCUUCGGGUGGAAUGUGGUACUCCCCAUUGGGCU ...(((((((...((((((.(((((((((.......)))))))))))))))...))(((((((.............)))))))..((((......)))).))))) ( -46.82) >consensus GUUGUUCCGCAAUGGGAGGUGGAGGCCGAAGCAGCAAAGCGCCAACUUCCUCGUACUCCACCU______CCUCCUCAGGUGGAAUGUGGUAUUCCCCAUUUAAGG ......(((((..(((((((...((((...........).))).))))))).....(((((((.............))))))).)))))................ (-19.46 = -18.77 + -0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:28 2006