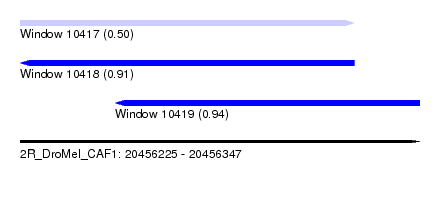
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,456,225 – 20,456,347 |
| Length | 122 |
| Max. P | 0.938006 |

| Location | 20,456,225 – 20,456,327 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 69.55 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -12.13 |
| Energy contribution | -11.69 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.45 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20456225 102 + 20766785 UACG-----------UAACUGCGCAUCCCGUCAUUAGCUUUUUUACUUUCGACGGCGACGGACGUUUCUUUUCAUCCGUGUCCGCGCAUCCUUAGUUGACUAAAUUCUGCCGC ...(-----------((..(((((.(((((((...((........))...))))).)).(((((..............))))))))))...((((....))))....)))... ( -23.24) >DroVir_CAF1 23506 101 + 1 ACAGUUGCGAUUUGGUGGCUGCGCAUUCCGUUAGUUGCUUUUUUCACUUCGGCGGCGGCAGUCGUUUCCUCU-------CGCCGCGCAGCGAACGCC-----AUUUUAGCCGC ...((.((((..((((((((((((...(((..(((..........))).))).(((((.((.......)).)-------)))))))))))...))))-----)..)).)).)) ( -33.80) >DroSec_CAF1 19723 98 + 1 UACG-----------UAACUGCGCAUUCCGUCAAUAGCUUUUUUACUUUUGACGACGACGGAUGUUUCCUUU----CGUGUCCGCGCAUCCUUAGUUCACUAAAUUCUGCCGC ...(-----------((..(((((....((((((.((........)).))))))((((.(((....)))..)----)))....)))))...((((....))))....)))... ( -23.60) >DroEre_CAF1 26490 102 + 1 UACG-----------UAACUGCGCAUUCCGCCAUUAGCUUUUUUACUUUCAACGGCGACGGAUGUUUCUUUUCAUUCGUGUCCGCGCAUCCUUAGUUCACUAAAUUCUGCGGC ..((-----------((..(((((....((((.....................))))(((((((........)))))))....)))))...((((....))))....)))).. ( -25.20) >DroYak_CAF1 19674 102 + 1 UACG-----------UAACUGCGCAUUCCGCCAUUAGCUUUUCUACUUUCGACGGCGACGGAUGUUUCUUUUCAUUCGUGUCCGCGCAUCCUUAGUUCACUAAAUUCUGCCGC ...(-----------((..(((((....((((.....................))))(((((((........)))))))....)))))...((((....))))....)))... ( -22.10) >DroMoj_CAF1 25931 101 + 1 UUCCUUACGAUUUGGUGGCUGCGCAUUCCGUUAGUUGCUUUUUUCACUUCAGCGGCGGCAGUCGCUCUCUUU-------CGCCGCGCAGCAAACGCC-----AUUUCAGCCGC ........((..(((((((((((((..........))).............(((((((.((.......)).)-------)))))))))))...))))-----)..))...... ( -34.20) >consensus UACG___________UAACUGCGCAUUCCGUCAUUAGCUUUUUUACUUUCGACGGCGACGGACGUUUCCUUU____CGUGUCCGCGCAUCCUUAGUUCACUAAAUUCUGCCGC ...................(((((....((((.....................))))..((((................)))))))))......................... (-12.13 = -11.69 + -0.44)

| Location | 20,456,225 – 20,456,327 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 69.55 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -13.74 |
| Energy contribution | -15.42 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20456225 102 - 20766785 GCGGCAGAAUUUAGUCAACUAAGGAUGCGCGGACACGGAUGAAAAGAAACGUCCGUCGCCGUCGAAAGUAAAAAAGCUAAUGACGGGAUGCGCAGUUA-----------CGUA ((((((...(((((....)))))..)))(((...(((((((........)))))))..((((((..(((......)))..))))))....))).....-----------))). ( -32.50) >DroVir_CAF1 23506 101 - 1 GCGGCUAAAAU-----GGCGUUCGCUGCGCGGCG-------AGAGGAAACGACUGCCGCCGCCGAAGUGAAAAAAGCAACUAACGGAAUGCGCAGCCACCAAAUCGCAACUGU ((((......(-----(((.((((((..((((((-------.(.(....).....)))))))...))))))....(((.(....)...)))...)))).....))))...... ( -33.80) >DroSec_CAF1 19723 98 - 1 GCGGCAGAAUUUAGUGAACUAAGGAUGCGCGGACACG----AAAGGAAACAUCCGUCGUCGUCAAAAGUAAAAAAGCUAUUGACGGAAUGCGCAGUUA-----------CGUA ((((((...(((((....)))))..)))(((((((((----...(....)...))).)))(((((.(((......))).)))))......))).....-----------))). ( -30.00) >DroEre_CAF1 26490 102 - 1 GCCGCAGAAUUUAGUGAACUAAGGAUGCGCGGACACGAAUGAAAAGAAACAUCCGUCGCCGUUGAAAGUAAAAAAGCUAAUGGCGGAAUGCGCAGUUA-----------CGUA ...(((...(((((....)))))..)))(((...(((.(((........))).)))(((((((...(((......)))))))))).....))).....-----------.... ( -25.20) >DroYak_CAF1 19674 102 - 1 GCGGCAGAAUUUAGUGAACUAAGGAUGCGCGGACACGAAUGAAAAGAAACAUCCGUCGCCGUCGAAAGUAGAAAAGCUAAUGGCGGAAUGCGCAGUUA-----------CGUA ((((((...(((((....)))))..)))(((...(((.(((........))).)))..((((((..(((......)))..))))))....))).....-----------))). ( -26.80) >DroMoj_CAF1 25931 101 - 1 GCGGCUGAAAU-----GGCGUUUGCUGCGCGGCG-------AAAGAGAGCGACUGCCGCCGCUGAAGUGAAAAAAGCAACUAACGGAAUGCGCAGCCACCAAAUCGUAAGGAA ..(((((....-----....((..((..((((((-------..((.......))..))))))...))..))....(((.(....)...))).)))))......((....)).. ( -30.10) >consensus GCGGCAGAAUUUAGUGAACUAAGGAUGCGCGGACACG____AAAAGAAACAUCCGUCGCCGUCGAAAGUAAAAAAGCUAAUGACGGAAUGCGCAGUUA___________CGUA ..(((....(((((....)))))..(((((((....................))....((((((..(((......)))..))))))...))))))))................ (-13.74 = -15.42 + 1.67)

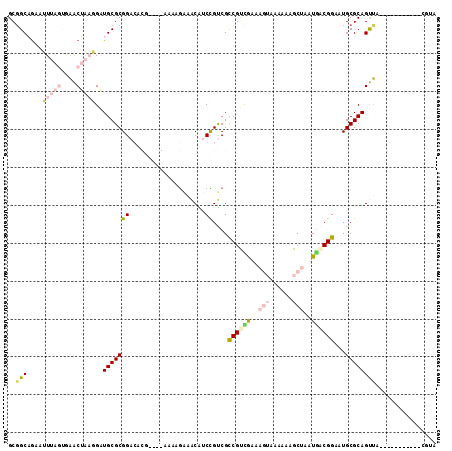
| Location | 20,456,254 – 20,456,347 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 67.75 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -11.36 |
| Energy contribution | -11.95 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
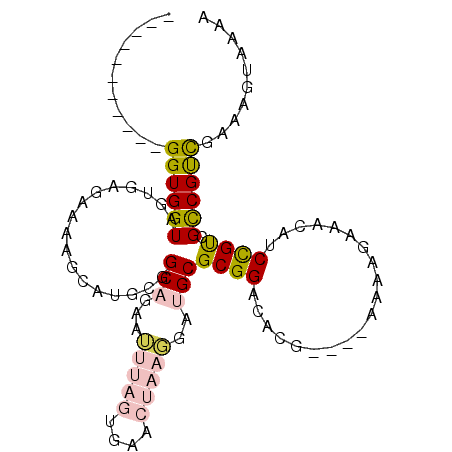
>2R_DroMel_CAF1 20456254 93 - 20766785 -----------GGUGGUAGUGAGAAAAGCAUGCGGCAGAAUUUAGUCAACUAAGGAUGCGCGGACACGGAUGAAAAGAAACGUCCGUCGCCGUCGAAAGUAAAA -----------((((............(..((((.((...(((((....)))))..))))))..)(((((((........)))))))))))..(....)..... ( -24.40) >DroVir_CAF1 23546 91 - 1 UUUUAUUUUUUGGUGGUAGCAAUA-AUGCGUGCGGCUAAAAU-----GGCGUUCGCUGCGCGGCG-------AGAGGAAACGACUGCCGCCGCCGAAGUGAAAA ((((((..(((((((((.(((...-.)))(((((((......-----.......)))))))((((-------...(....)...))))))))))))))))))). ( -35.32) >DroSec_CAF1 19752 89 - 1 -----------GGUGGUAGUGAGAAAAGCAUGCGGCAGAAUUUAGUGAACUAAGGAUGCGCGGACACG----AAAGGAAACAUCCGUCGUCGUCAAAAGUAAAA -----------................((.((((.((...(((((....)))))..))))))((((((----(..(((....))).)))).)))....)).... ( -20.50) >DroEre_CAF1 26519 93 - 1 -----------GGUGGUAGUGAGAGAAGCAUGCCGCAGAAUUUAGUGAACUAAGGAUGCGCGGACACGAAUGAAAAGAAACAUCCGUCGCCGUUGAAAGUAAAA -----------...((((((.......)).))))(((...(((((....)))))..)))((((..(((.(((........))).)))..))))........... ( -21.50) >DroYak_CAF1 19703 93 - 1 -----------GGUGGUACUGAGAAAAGCAUGCGGCAGAAUUUAGUGAACUAAGGAUGCGCGGACACGAAUGAAAAGAAACAUCCGUCGCCGUCGAAAGUAGAA -----------.....((((..((...((.....(((...(((((....)))))..)))(((((......(....)......))))).))..))...))))... ( -19.80) >DroMoj_CAF1 25971 91 - 1 UUUUAUUUUUUGGUGGUAGCAAUA-AUAUGUGCGGCUGAAAU-----GGCGUUUGCUGCGCGGCG-------AAAGAGAGCGACUGCCGCCGCUGAAGUGAAAA (((((((((.((((((((((....-...((((((((......-----.......))))))))((.-------.......))).)))))))))..))))))))). ( -33.42) >consensus ___________GGUGGUAGUGAGAAAAGCAUGCGGCAGAAUUUAGUGAACUAAGGAUGCGCGGACACG____AAAAGAAACAUCCGUCGCCGUCGAAAGUAAAA ...........((((((.................(((...(((((....)))))..)))((((....................)))).)))))).......... (-11.36 = -11.95 + 0.59)

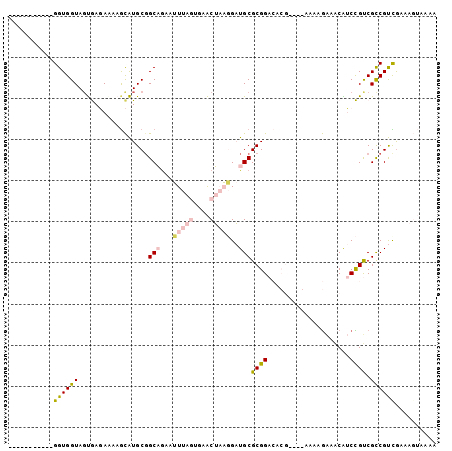
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:25 2006