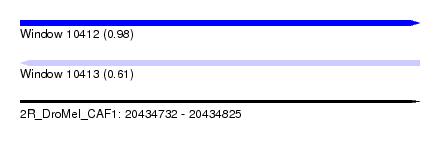
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,434,732 – 20,434,825 |
| Length | 93 |
| Max. P | 0.983361 |

| Location | 20,434,732 – 20,434,825 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 64.74 |
| Mean single sequence MFE | -24.05 |
| Consensus MFE | -12.67 |
| Energy contribution | -13.27 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
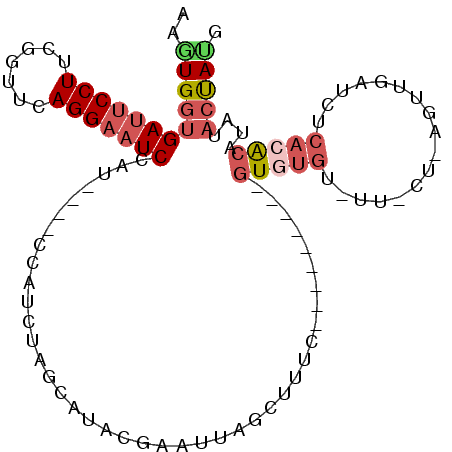
>2R_DroMel_CAF1 20434732 93 + 20766785 CGUAGUACAUGUGUGAGAUCAACUCAGAAACACACAC---------GAAAGCUAAUUCGUGUGCUAGAUGG----ACGGAUUCCUGAACCGAAGGAAUCACCACCU ..((((((..(((((...((......))..)))))((---------(((......)))))))))))..(((----...(((((((.......))))))).)))... ( -27.60) >DroSec_CAF1 44138 87 + 1 CAUAGUAUAUGUGUGAGAUCAA----------CACAC---------GAAAGCUAAUUCGUAUGCUAGAUGGAUGGAUGGAUUCCUGAACCGAAGGAAUCACCACUU ..(((((((((((((.......----------)))))---------(((......)))))))))))......(((...(((((((.......))))))).)))... ( -24.70) >DroSim_CAF1 44575 87 + 1 CAUAGUAUAUGUGUGAGAUCAA----------CACAC---------GAAGGCUAAUUCGUAUGCUAGAUGGAUGGAUGGAUUCCUGAACCGAAGGAAUCACCACUU ..(((((((((((((.......----------)))))---------(((......)))))))))))......(((...(((((((.......))))))).)))... ( -25.50) >DroYak_CAF1 45002 93 + 1 CAUAGUACAUGUGUGAGAUCAACUUAGGAAUACAAAC---------AAAAACCAAUUCGUGUGCUAGAUGA----AUGGAUUCCUGAACUGGAGGAAUCACCAUUU ..(((((((((..((((.....))))((.........---------.....))....)))))))))...((----((((((((((.......))))))..)))))) ( -20.24) >DroAna_CAF1 40387 90 + 1 GUUGGC-UAUUCG------GAGCUGAGGAAUAUUAAUUGGCCCGCUGAGAACCAAUUCUCAUCCCAAC--------AAGAGACCUAAACCAAAGGAAUCUACAGC- (((((.-....((------(.((..(..........)..))))).((((((....))))))..)))))--------.(((..(((.......)))..))).....- ( -22.20) >consensus CAUAGUAUAUGUGUGAGAUCAACU_AG_AA_ACACAC_________GAAAGCUAAUUCGUAUGCUAGAUGG____AUGGAUUCCUGAACCGAAGGAAUCACCACUU ..(((((((((..............................................)))))))))............(((((((.......)))))))....... (-12.67 = -13.27 + 0.60)


| Location | 20,434,732 – 20,434,825 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 64.74 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -7.99 |
| Energy contribution | -9.43 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20434732 93 - 20766785 AGGUGGUGAUUCCUUCGGUUCAGGAAUCCGU----CCAUCUAGCACACGAAUUAGCUUUC---------GUGUGUGUUUCUGAGUUGAUCUCACACAUGUACUACG ((((((.(((((((.......)))))))...----)))))).(((((((((......)))---------)))))).....((((.....))))............. ( -30.60) >DroSec_CAF1 44138 87 - 1 AAGUGGUGAUUCCUUCGGUUCAGGAAUCCAUCCAUCCAUCUAGCAUACGAAUUAGCUUUC---------GUGUG----------UUGAUCUCACACAUAUACUAUG ..((((.(((((((.......)))))))...))))..(((.((((((((((......)))---------)))))----------)))))................. ( -24.00) >DroSim_CAF1 44575 87 - 1 AAGUGGUGAUUCCUUCGGUUCAGGAAUCCAUCCAUCCAUCUAGCAUACGAAUUAGCCUUC---------GUGUG----------UUGAUCUCACACAUAUACUAUG ..((((.(((((((.......)))))))...))))..(((.((((((((((......)))---------)))))----------)))))................. ( -23.20) >DroYak_CAF1 45002 93 - 1 AAAUGGUGAUUCCUCCAGUUCAGGAAUCCAU----UCAUCUAGCACACGAAUUGGUUUUU---------GUUUGUAUUCCUAAGUUGAUCUCACACAUGUACUAUG ..(((((((......((..(.((((((.((.----.((...(((.((.....)))))..)---------)..)).)))))).)..))...)))).)))........ ( -15.70) >DroAna_CAF1 40387 90 - 1 -GCUGUAGAUUCCUUUGGUUUAGGUCUCUU--------GUUGGGAUGAGAAUUGGUUCUCAGCGGGCCAAUUAAUAUUCCUCAGCUC------CGAAUA-GCCAAC -((((((((..(((.......)))..))).--------(((((((....(((((((((.....))))))))).....))).))))..------...)))-)).... ( -26.10) >consensus AAGUGGUGAUUCCUUCGGUUCAGGAAUCCAU____CCAUCUAGCAUACGAAUUAGCUUUC_________GUGUGU_UU_CU_AGUUGAUCUCACACAUAUACUAUG ..((((((((((((.......))))))).........................................(((((.................)))))....))))). ( -7.99 = -9.43 + 1.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:20 2006