
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,431,141 – 20,431,261 |
| Length | 120 |
| Max. P | 0.522860 |

| Location | 20,431,141 – 20,431,261 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.56 |
| Mean single sequence MFE | -40.13 |
| Consensus MFE | -26.67 |
| Energy contribution | -28.48 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20431141 120 + 20766785 CCAUCAUACGGUGGCUCAUCCAGACGGCCGAUUACUGCAUUUAGUAGCUUGGAGUCUGGAGUAAGUCGCAGGUUUGGCUCCUUAAGCAGUCGCUCCAACGCCUGCCAGUUACCGUAUAUC .....(((((((((((..(((((((..((((((((((....)))))).)))).))))))).......((((((((((.......(((....))))))).)))))).)))))))))))... ( -48.41) >DroGri_CAF1 50294 120 + 1 UCAUCCAGCGGCGGUUCCUCCAAUCUGCUGAUCACUGUAUUCAGUAGCGAAGAGCGUGGAGUCAGCUUUAGCUCCGGAUGCUUCAGCAGUUUCUCCAGAGCACGCCAAUUGCCCCAAAUG .........((((((((.......(((((((.....((((((....((.....))..(((((........))))))))))).)))))))........)))).)))).............. ( -37.56) >DroSec_CAF1 40534 120 + 1 CUAUCAUACGGUGGCUCAUCCAGACGGCCGAUCACUGCAUUUAGUAGCUUAGAGUCUGGAGUAAGUCGCAGGUUUGGCUCCUUCAGAAGUCGCUCCAACGCCUGCCAGUUACCGUAUAUC .....(((((((((((..(((((((..(..(...((((.....)))).)..).))))))).......(((((((((((..((.....))..))..))).)))))).)))))))))))... ( -39.60) >DroSim_CAF1 40955 120 + 1 CCAUCAUACGGUGGCUCAUCCAGACGGCCGAUCACUGCAUUUAGUAGCUUAGAGUCUGGAGUAAGUCGCAGGUUUGGCUCCUUCAGAAGUCGCUCCAACGCCUGCCAGUUACCGUAUAUC .....(((((((((((..(((((((..(..(...((((.....)))).)..).))))))).......(((((((((((..((.....))..))..))).)))))).)))))))))))... ( -39.60) >DroEre_CAF1 41276 120 + 1 CCAUCGAAUGGCGGCUCAACCAAACGACCCAUCACUGCAUUUAGUAGCUUGGACUCUGCAGUAAGGCGCAGGCUUGGCUCCUUCAGCAGUCGCUCCAAGGCCUGCCAGUUACCGUAUAUC ........(((((((...................((((.....))))((((((..((((.(.((((.((.......)).))))).))))....))))))))).))))............. ( -33.30) >DroYak_CAF1 41403 120 + 1 CCAUCGUACGGCGGCUCAUCCAGUCGGCCGAUCACCGAAUUUAGUAGCUUGGAGUCUGGAGCGAGUCGCAGACUUGGCUCCUUCAGCAGUCGCUCCAACGCCUGCCAGUUACCGUAUAUC .....((((((..(((.......((((.......)))).....((((((((((((((((((.((((((......)))))))))))).....))))))).).)))).)))..))))))... ( -42.30) >consensus CCAUCAUACGGCGGCUCAUCCAGACGGCCGAUCACUGCAUUUAGUAGCUUAGAGUCUGGAGUAAGUCGCAGGUUUGGCUCCUUCAGCAGUCGCUCCAACGCCUGCCAGUUACCGUAUAUC .....(((((((((((..(((((((..((((.(((((....)))).).)))).))))))).......(((((((((((..((.....))..))..))).)))))).)))))))))))... (-26.67 = -28.48 + 1.81)

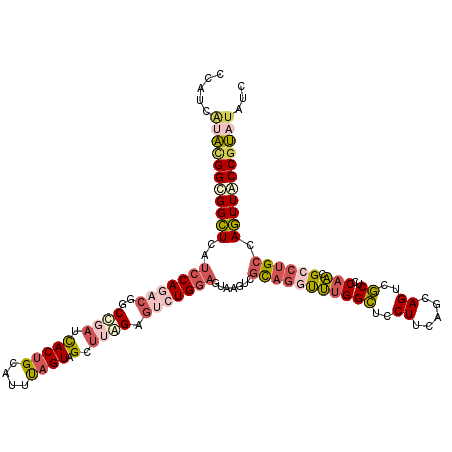
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:16 2006