
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,426,280 – 20,426,388 |
| Length | 108 |
| Max. P | 0.996833 |

| Location | 20,426,280 – 20,426,388 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 93.64 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -34.63 |
| Energy contribution | -34.36 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
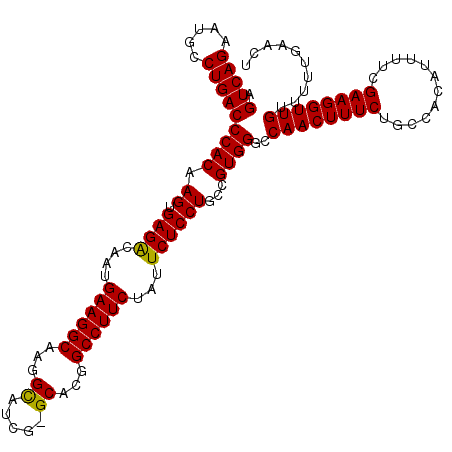
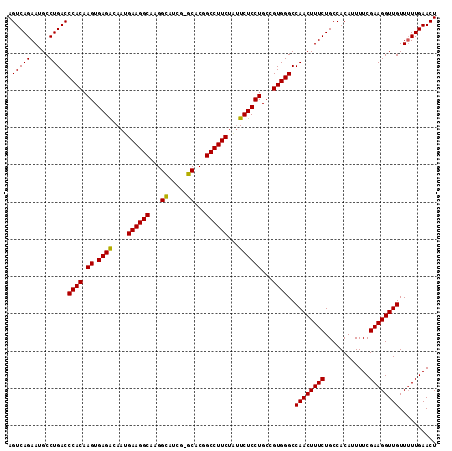
>2R_DroMel_CAF1 20426280 108 + 20766785 AGUCAGAAUGCCUGACCCACAAGUGAGGCAAUGAAGGCAAGG-------CACGGCCUUCUAUUCUCCUGCCGUGGGCCAACUUUCUGCCACAUUUUCGAAGGUUGUUUUUGAACU ..((((((.......(((((.((.((((....((((((....-------....))))))...))))))...))))).((((((((............)))))))).))))))... ( -32.60) >DroSec_CAF1 35695 114 + 1 AGUCAGAAUGCCUGACCCACAAGUGAGACAAUGAAGGCAAGGCAUCG-GCACGGCCUUCUAUUCUCCUGCCGUGGGCCAACUUUCUGCCACAUUUUCGAAGGUUGUUUUUGAACU ..((((((.......(((((.((.((((....((((((...((....-))...))))))...))))))...))))).((((((((............)))))))).))))))... ( -37.10) >DroSim_CAF1 36079 114 + 1 AGUCAGAAUGCCUGACCCACAAGUGAGACAAUGAAGGCAAGGCAUCG-GCACGGCCUUCUAUUCUCCUGCCGUGGGCCAACUUUCUGCCACAUUUUCGAAGGUUGUUUUUGAACU ..((((((.......(((((.((.((((....((((((...((....-))...))))))...))))))...))))).((((((((............)))))))).))))))... ( -37.10) >DroEre_CAF1 36363 114 + 1 AGUCAGAAUGCCUGACCCACAAGUGAGACAAUGAAGGCAAGGUGUUGUGCACGGCCUUCUCUUCUCCUGCCGUGGGCCAACUUUCUGCCACAUUUUCGAAGGUUGUU-UUGAACU ..((((((.......(((((.((.((((....((((((...(((.....))).))))))...))))))...))))).((((((((............))))))))))-))))... ( -37.00) >DroYak_CAF1 36390 115 + 1 AGUCAGAAAGCCUGACCCACAAGUGAGACAAUGAAGGCGAGGUGUUGUGCACGGCCUUCAGCUCUCCUGCCGUGGGCCAACUUUCUGCCACAUUUUCGAAGGUUGUUUUUGAACU .(.(((((((..((.(((((.((.((((...(((((((...(((.....))).)))))))..))))))...))))).)).))))))).).....(((((((.....))))))).. ( -40.20) >consensus AGUCAGAAUGCCUGACCCACAAGUGAGACAAUGAAGGCAAGGCAUCG_GCACGGCCUUCUAUUCUCCUGCCGUGGGCCAACUUUCUGCCACAUUUUCGAAGGUUGUUUUUGAACU .(((((.....)))))((((.((.((((....((((((...((.....))...))))))...))))))...))))..((((((((............)))))))).......... (-34.63 = -34.36 + -0.27)

| Location | 20,426,280 – 20,426,388 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 93.64 |
| Mean single sequence MFE | -37.78 |
| Consensus MFE | -34.34 |
| Energy contribution | -34.62 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
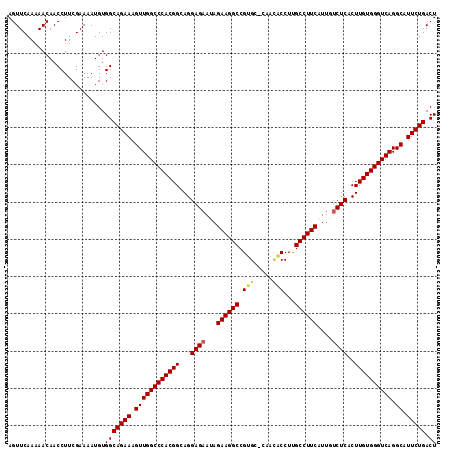
>2R_DroMel_CAF1 20426280 108 - 20766785 AGUUCAAAAACAACCUUCGAAAAUGUGGCAGAAAGUUGGCCCACGGCAGGAGAAUAGAAGGCCGUG-------CCUUGCCUUCAUUGCCUCACUUGUGGGUCAGGCAUUCUGACU ..........................(((((((.((((((((((((.(((..(((.((((((....-------....))))))))).)))...)))))))))).)).))))).)) ( -33.30) >DroSec_CAF1 35695 114 - 1 AGUUCAAAAACAACCUUCGAAAAUGUGGCAGAAAGUUGGCCCACGGCAGGAGAAUAGAAGGCCGUGC-CGAUGCCUUGCCUUCAUUGUCUCACUUGUGGGUCAGGCAUUCUGACU ..........................(((((((.((((((((((((...((((...((((((...((-....))...))))))....))))..)))))))))).)).))))).)) ( -37.40) >DroSim_CAF1 36079 114 - 1 AGUUCAAAAACAACCUUCGAAAAUGUGGCAGAAAGUUGGCCCACGGCAGGAGAAUAGAAGGCCGUGC-CGAUGCCUUGCCUUCAUUGUCUCACUUGUGGGUCAGGCAUUCUGACU ..........................(((((((.((((((((((((...((((...((((((...((-....))...))))))....))))..)))))))))).)).))))).)) ( -37.40) >DroEre_CAF1 36363 114 - 1 AGUUCAA-AACAACCUUCGAAAAUGUGGCAGAAAGUUGGCCCACGGCAGGAGAAGAGAAGGCCGUGCACAACACCUUGCCUUCAUUGUCUCACUUGUGGGUCAGGCAUUCUGACU .......-..................(((((((.((((((((((((...((((.((((((((.(((.....)))...)))))).)).))))..)))))))))).)).))))).)) ( -38.10) >DroYak_CAF1 36390 115 - 1 AGUUCAAAAACAACCUUCGAAAAUGUGGCAGAAAGUUGGCCCACGGCAGGAGAGCUGAAGGCCGUGCACAACACCUCGCCUUCAUUGUCUCACUUGUGGGUCAGGCUUUCUGACU ..........................((((((((((((((((((((...((((..(((((((.(((.....)))...)))))))...))))..)))))))))).)))))))).)) ( -42.70) >consensus AGUUCAAAAACAACCUUCGAAAAUGUGGCAGAAAGUUGGCCCACGGCAGGAGAAUAGAAGGCCGUGC_CAACACCUUGCCUUCAUUGUCUCACUUGUGGGUCAGGCAUUCUGACU ..........................(((((((.((((((((((((...((((...((((((.(((.....)))...))))))....))))..)))))))))).)).))))).)) (-34.34 = -34.62 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:15 2006