
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,422,270 – 20,422,590 |
| Length | 320 |
| Max. P | 0.999990 |

| Location | 20,422,270 – 20,422,390 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.22 |
| Mean single sequence MFE | -25.16 |
| Consensus MFE | -23.91 |
| Energy contribution | -23.24 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
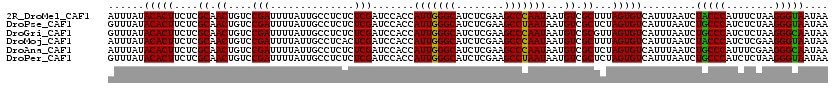
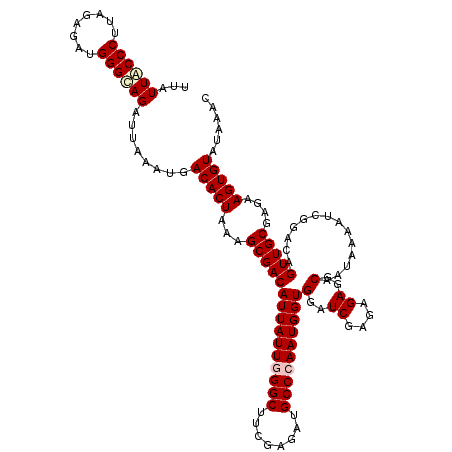
>2R_DroMel_CAF1 20422270 120 + 20766785 AUUUAUACACUUCUCGCAACUGUCCGAUUUUAUUGCCUCUCUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUACCCAUUUCUAAGGGUAAUAA ......(((((....((.((....(((..............))).......(((((((........)))))))...)).))...))))).........(((((........))))).... ( -23.34) >DroPse_CAF1 35654 120 + 1 GUUUAUACACUUCUCGCAACUGUCCGAUUUUAUUGCCUCUCUCGAUCCACCAUUGGGCAUCUCGAAGCCUAAUAAUGUCGCUCUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGUAAUAA .............(((........)))..(((((((((.....(((.....(((((((........)))))))((((.(((....))).))))..........))).....))))))))) ( -21.30) >DroGri_CAF1 39665 120 + 1 GUUUAUACACUUCUCGCAACUGUCCGAUUUUAUUGCCUCUCUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCGUUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGCAAUAA ......(((((...(((.((....(((..............))).......(((((((........)))))))...)).)))..))))).........(((((........))))).... ( -28.84) >DroMoj_CAF1 43904 120 + 1 AUUUAUACACUUCUCGCAACUGUCCGAUUUUAUUGCCUCACUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUACCCAUCUCGAAGGGUAAUAA .............(((........)))..(((((((((...((((......(((((((........)))))))((((.(((....))).)))).............)))).))))))))) ( -26.70) >DroAna_CAF1 27677 120 + 1 AUUUAUACACUUCUCGCAACUGUCCGAUUUUAUUGCCUCUCUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUCUAGUGUCAUUUAAUCUGCCCAUUUCGAAGGGCAAUAA .............(((........)))..((((((((.((.((((......(((((((........)))))))((((.(((....))).)))).............)))))))))))))) ( -29.50) >DroPer_CAF1 35428 120 + 1 GUUUAUACACUUCUCGCAACUGUCCGAUUUUAUUGCCUCUCUCGAUCCACCAUUGGGCAUCUCGAAGCCUAAUAAUGUCGCUCUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGUAAUAA .............(((........)))..(((((((((.....(((.....(((((((........)))))))((((.(((....))).))))..........))).....))))))))) ( -21.30) >consensus AUUUAUACACUUCUCGCAACUGUCCGAUUUUAUUGCCUCUCUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUCUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGUAAUAA ......(((((....((.((....(((..............))).......(((((((........)))))))...)).))...))))).........(((((........))))).... (-23.91 = -23.24 + -0.67)

| Location | 20,422,270 – 20,422,390 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.22 |
| Mean single sequence MFE | -33.64 |
| Consensus MFE | -32.85 |
| Energy contribution | -33.30 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.30 |
| SVM RNA-class probability | 0.999865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
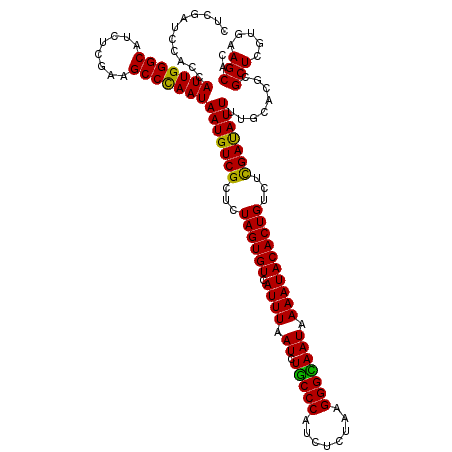
>2R_DroMel_CAF1 20422270 120 - 20766785 UUAUUACCCUUAGAAAUGGGUAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAGAGAGGCAAUAAAAUCGGACAGUUGCGAGAAGUGUAUAAAU ...((((((........))))))........(((((...(((((((((((((((........))))))))))(..((....))..)..............)))))....)))))...... ( -36.30) >DroPse_CAF1 35654 120 - 1 UUAUUACCCUUAGAGAUGGGCAGAUUAAAUGACACUAGAGCGACAUUAUUAGGCUUCGAGAUGCCCAAUGGUGGAUCGAGAGAGGCAAUAAAAUCGGACAGUUGCGAGAAGUGUAUAAAC ......(((........)))...........(((((...(((((((((((.(((........))).))))))(..((....))..)..............)))))....)))))...... ( -26.00) >DroGri_CAF1 39665 120 - 1 UUAUUGCCCUUAGAGAUGGGCAGAUUAAAUGACACUAACGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAGAGAGGCAAUAAAAUCGGACAGUUGCGAGAAGUGUAUAAAC ...((((((........))))))........(((((..((((((((((((((((........))))))))))(..((....))..)..............))))))...)))))...... ( -40.00) >DroMoj_CAF1 43904 120 - 1 UUAUUACCCUUCGAGAUGGGUAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAGUGAGGCAAUAAAAUCGGACAGUUGCGAGAAGUGUAUAAAU ...((((((........))))))........(((((...(((((((((((((((........))))))))))...((((..............))))...)))))....)))))...... ( -35.24) >DroAna_CAF1 27677 120 - 1 UUAUUGCCCUUCGAAAUGGGCAGAUUAAAUGACACUAGAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAGAGAGGCAAUAAAAUCGGACAGUUGCGAGAAGUGUAUAAAU ...((((((........))))))........(((((...(((((((((((((((........))))))))))(..((....))..)..............)))))....)))))...... ( -38.30) >DroPer_CAF1 35428 120 - 1 UUAUUACCCUUAGAGAUGGGCAGAUUAAAUGACACUAGAGCGACAUUAUUAGGCUUCGAGAUGCCCAAUGGUGGAUCGAGAGAGGCAAUAAAAUCGGACAGUUGCGAGAAGUGUAUAAAC ......(((........)))...........(((((...(((((((((((.(((........))).))))))(..((....))..)..............)))))....)))))...... ( -26.00) >consensus UUAUUACCCUUAGAGAUGGGCAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAGAGAGGCAAUAAAAUCGGACAGUUGCGAGAAGUGUAUAAAC ...((((((........))))))........(((((...(((((((((((((((........))))))))))(..((....))..)..............)))))....)))))...... (-32.85 = -33.30 + 0.45)


| Location | 20,422,310 – 20,422,430 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.06 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -33.54 |
| Energy contribution | -32.57 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.71 |
| Structure conservation index | 1.00 |
| SVM decision value | 5.57 |
| SVM RNA-class probability | 0.999990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20422310 120 + 20766785 CUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUACCCAUUUCUAAGGGUAAUAAAAUACACUGUCUCGACAUUUUGCACGCGCUCGUUAAGCAC ...........(((((((........)))))))(((((((...((((((.((((.((.(((((........))))))).))))))))))...)))))))........(((.....))).. ( -34.00) >DroPse_CAF1 35694 120 + 1 CUCGAUCCACCAUUGGGCAUCUCGAAGCCUAAUAAUGUCGCUCUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGUAAUAAAAUACACUGUCUCGAUAUUUUGCACGCGCUCGUGAAGCAC ......(((....)))((.((.(((.((.....(((((((...((((((.((((.((.(((((........))))))).))))))))))...)))))))......))..))).)).)).. ( -30.80) >DroGri_CAF1 39705 120 + 1 CUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCGUUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGCAAUAAAAUACACUGUCUUGACAUUUUGCACGCGCUAGUUAAGCAC ...........(((((((........)))))))(((((((.(.((((((.((((.((.(((((........))))))).)))))))))).).)))))))........(((.....))).. ( -35.10) >DroWil_CAF1 44294 120 + 1 CUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUUUAGUGUCAUUUAAUCUGCCCAUUUCGAGGGGCAAUAAAAUACACUGUCUCGAUAUUUUGCACGCGCUGGUUAAGCAC ...........(((((((........)))))))(((((((...((((((.((((.((.(((((........))))))).))))))))))...)))))))........(((.....))).. ( -35.20) >DroAna_CAF1 27717 120 + 1 CUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUCUAGUGUCAUUUAAUCUGCCCAUUUCGAAGGGCAAUAAAAUACACUGUCUCGAUAUUUUGCACGCGCUCGUGAAGCAC ...........(((((((........)))))))(((((((...((((((.((((.((.(((((........))))))).))))))))))...))))))).(((.(((....)))..))). ( -35.00) >DroPer_CAF1 35468 120 + 1 CUCGAUCCACCAUUGGGCAUCUCGAAGCCUAAUAAUGUCGCUCUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGUAAUAAAAUACACUGUCUCGAUAUUUUGCACGCGCUCGUGAAGCAC ......(((....)))((.((.(((.((.....(((((((...((((((.((((.((.(((((........))))))).))))))))))...)))))))......))..))).)).)).. ( -30.80) >consensus CUCGAUCCACCAUUGGGCAUCUCGAAGCCCAAUAAUGUCGCUCUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGCAAUAAAAUACACUGUCUCGAUAUUUUGCACGCGCUCGUGAAGCAC ...........(((((((........)))))))(((((((...((((((.((((.((.(((((........))))))).))))))))))...)))))))........(((.....))).. (-33.54 = -32.57 + -0.97)


| Location | 20,422,310 – 20,422,430 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.06 |
| Mean single sequence MFE | -38.18 |
| Consensus MFE | -37.02 |
| Energy contribution | -37.68 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.24 |
| SVM RNA-class probability | 0.999847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20422310 120 - 20766785 GUGCUUAACGAGCGCGUGCAAAAUGUCGAGACAGUGUAUUUUAUUACCCUUAGAAAUGGGUAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAG ((((((...))))))((.((((((((((....(((((((((..((((((........))))))...)))).)))))....))))))).))).)).((((..((((....))))..)))). ( -41.70) >DroPse_CAF1 35694 120 - 1 GUGCUUCACGAGCGCGUGCAAAAUAUCGAGACAGUGUAUUUUAUUACCCUUAGAGAUGGGCAGAUUAAAUGACACUAGAGCGACAUUAUUAGGCUUCGAGAUGCCCAAUGGUGGAUCGAG ((((((...))))))..........((((...(((((((((.(((.(((........)))..))).)))).))))).......(((((((.(((........))).)))))))..)))). ( -31.00) >DroGri_CAF1 39705 120 - 1 GUGCUUAACUAGCGCGUGCAAAAUGUCAAGACAGUGUAUUUUAUUGCCCUUAGAGAUGGGCAGAUUAAAUGACACUAACGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAG (((((.....))))).(((.....((....))(((((((((..((((((........))))))...)))).)))))...))).(((((((((((........)))))))))))....... ( -41.40) >DroWil_CAF1 44294 120 - 1 GUGCUUAACCAGCGCGUGCAAAAUAUCGAGACAGUGUAUUUUAUUGCCCCUCGAAAUGGGCAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAG (((((.....)))))..........((((...(((((((((..((((((........))))))...)))).))))).......(((((((((((........)))))))))))..)))). ( -41.10) >DroAna_CAF1 27717 120 - 1 GUGCUUCACGAGCGCGUGCAAAAUAUCGAGACAGUGUAUUUUAUUGCCCUUCGAAAUGGGCAGAUUAAAUGACACUAGAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAG ((((((...))))))..........((((...(((((((((..((((((........))))))...)))).))))).......(((((((((((........)))))))))))..)))). ( -42.90) >DroPer_CAF1 35468 120 - 1 GUGCUUCACGAGCGCGUGCAAAAUAUCGAGACAGUGUAUUUUAUUACCCUUAGAGAUGGGCAGAUUAAAUGACACUAGAGCGACAUUAUUAGGCUUCGAGAUGCCCAAUGGUGGAUCGAG ((((((...))))))..........((((...(((((((((.(((.(((........)))..))).)))).))))).......(((((((.(((........))).)))))))..)))). ( -31.00) >consensus GUGCUUAACGAGCGCGUGCAAAAUAUCGAGACAGUGUAUUUUAUUACCCUUAGAAAUGGGCAGAUUAAAUGACACUAAAGCGACAUUAUUGGGCUUCGAGAUGCCCAAUGGUGGAUCGAG (((((.....)))))..........((((...(((((((((..((((((........))))))...)))).))))).......(((((((((((........)))))))))))..)))). (-37.02 = -37.68 + 0.67)

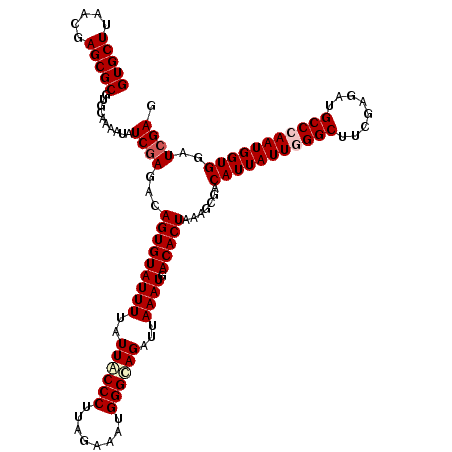
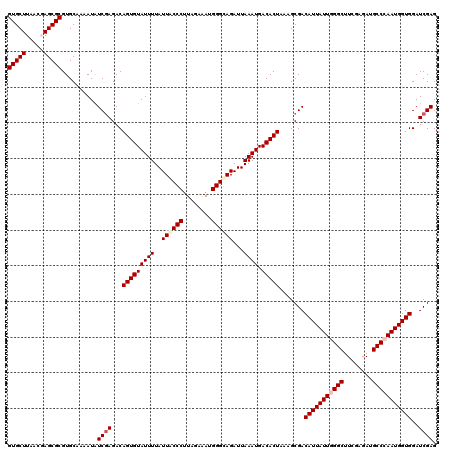
| Location | 20,422,350 – 20,422,470 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.22 |
| Mean single sequence MFE | -36.63 |
| Consensus MFE | -36.39 |
| Energy contribution | -35.50 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.53 |
| SVM RNA-class probability | 0.999989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20422350 120 + 20766785 CUUUAGUGUCAUUUAAUCUACCCAUUUCUAAGGGUAAUAAAAUACACUGUCUCGACAUUUUGCACGCGCUCGUUAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGU ...((((((.((((.((.(((((........))))))).))))))))))(((((((((...(((((.(((.....))).)))))......)))))))))..................... ( -36.20) >DroPse_CAF1 35734 120 + 1 CUCUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGUAAUAAAAUACACUGUCUCGAUAUUUUGCACGCGCUCGUGAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGU (((((((((.((((.((.(((((........))))))).)))))))))).((((((((...(((((.(((.....))).)))))......)))))))))))................... ( -35.80) >DroGri_CAF1 39745 120 + 1 CGUUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGCAAUAAAAUACACUGUCUUGACAUUUUGCACGCGCUAGUUAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGU .(.((((((.((((.((.(((((........))))))).)))))))))).).((((.....(((((.(((.....))).)))))...))))(((((.(....))))))............ ( -38.40) >DroWil_CAF1 44334 120 + 1 CUUUAGUGUCAUUUAAUCUGCCCAUUUCGAGGGGCAAUAAAAUACACUGUCUCGAUAUUUUGCACGCGCUGGUUAAGCACGUGUUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGU ...((((((.((((.((.(((((........))))))).))))))))))(((((((((...(((((.(((.....))).)))))......)))))))))..................... ( -35.80) >DroAna_CAF1 27757 120 + 1 CUCUAGUGUCAUUUAAUCUGCCCAUUUCGAAGGGCAAUAAAAUACACUGUCUCGAUAUUUUGCACGCGCUCGUGAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGU (((((((((.((((.((.(((((........))))))).)))))))))).((((((((...(((((.(((.....))).)))))......)))))))))))................... ( -37.80) >DroPer_CAF1 35508 120 + 1 CUCUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGUAAUAAAAUACACUGUCUCGAUAUUUUGCACGCGCUCGUGAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGU (((((((((.((((.((.(((((........))))))).)))))))))).((((((((...(((((.(((.....))).)))))......)))))))))))................... ( -35.80) >consensus CUCUAGUGUCAUUUAAUCUGCCCAUCUCUAAGGGCAAUAAAAUACACUGUCUCGAUAUUUUGCACGCGCUCGUGAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGU ...((((((.((((.((.(((((........))))))).))))))))))(((((((((...(((((.(((.....))).)))))......)))))))))..................... (-36.39 = -35.50 + -0.89)

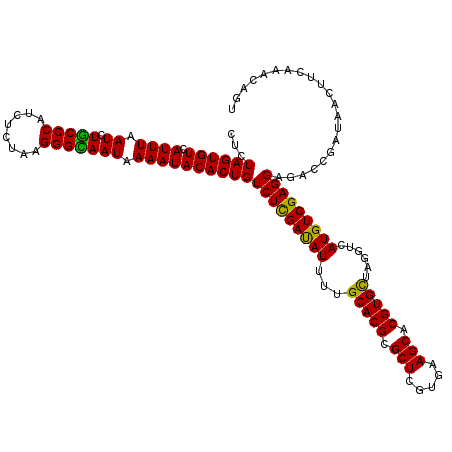
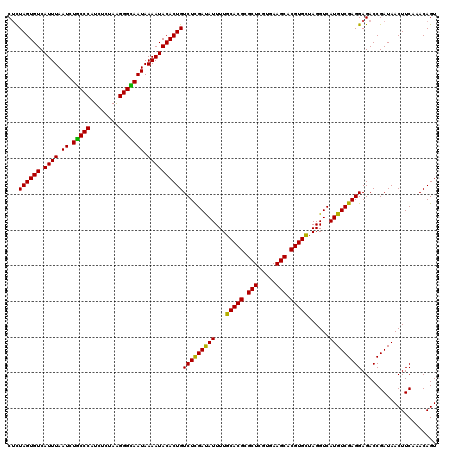
| Location | 20,422,350 – 20,422,470 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.22 |
| Mean single sequence MFE | -37.35 |
| Consensus MFE | -36.12 |
| Energy contribution | -36.45 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.80 |
| SVM RNA-class probability | 0.999952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20422350 120 - 20766785 ACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUAACGAGCGCGUGCAAAAUGUCGAGACAGUGUAUUUUAUUACCCUUAGAAAUGGGUAGAUUAAAUGACACUAAAG ((((...........))))...((((((((......((((((((((...))))))))))...))))))))..(((((((((..((((((........))))))...)))).))))).... ( -41.00) >DroPse_CAF1 35734 120 - 1 ACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUCACGAGCGCGUGCAAAAUAUCGAGACAGUGUAUUUUAUUACCCUUAGAGAUGGGCAGAUUAAAUGACACUAGAG ...((((((.(((...((((..........))))..((((((((((...))))))))))..))).)).))))(((((((((.(((.(((........)))..))).)))).))))).... ( -34.60) >DroGri_CAF1 39745 120 - 1 ACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUAACUAGCGCGUGCAAAAUGUCAAGACAGUGUAUUUUAUUGCCCUUAGAGAUGGGCAGAUUAAAUGACACUAACG ....(((((.(((...((((..........))))..(((((((((.....)))))))))..))).)))))..(((((((((..((((((........))))))...)))).))))).... ( -40.10) >DroWil_CAF1 44334 120 - 1 ACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAACACGUGCUUAACCAGCGCGUGCAAAAUAUCGAGACAGUGUAUUUUAUUGCCCCUCGAAAUGGGCAGAUUAAAUGACACUAAAG ...((((((.(((...((((..........))))...((((((((.....))))))))...))).)).))))(((((((((..((((((........))))))...)))).))))).... ( -33.80) >DroAna_CAF1 27757 120 - 1 ACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUCACGAGCGCGUGCAAAAUAUCGAGACAGUGUAUUUUAUUGCCCUUCGAAAUGGGCAGAUUAAAUGACACUAGAG ...((((((.(((...((((..........))))..((((((((((...))))))))))..))).)).))))(((((((((..((((((........))))))...)))).))))).... ( -40.00) >DroPer_CAF1 35508 120 - 1 ACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUCACGAGCGCGUGCAAAAUAUCGAGACAGUGUAUUUUAUUACCCUUAGAGAUGGGCAGAUUAAAUGACACUAGAG ...((((((.(((...((((..........))))..((((((((((...))))))))))..))).)).))))(((((((((.(((.(((........)))..))).)))).))))).... ( -34.60) >consensus ACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUAACGAGCGCGUGCAAAAUAUCGAGACAGUGUAUUUUAUUACCCUUAGAAAUGGGCAGAUUAAAUGACACUAAAG ...((((((.(((...((((..........))))..(((((((((.....)))))))))..))).)).))))(((((((((..((((((........))))))...)))).))))).... (-36.12 = -36.45 + 0.33)
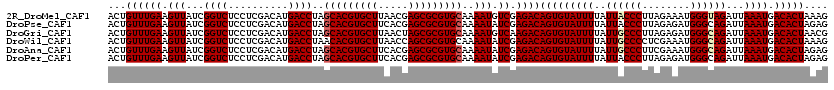
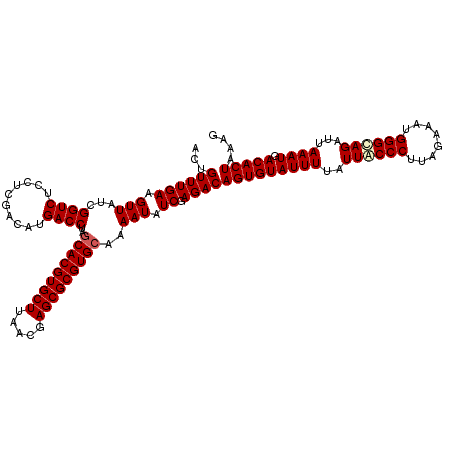
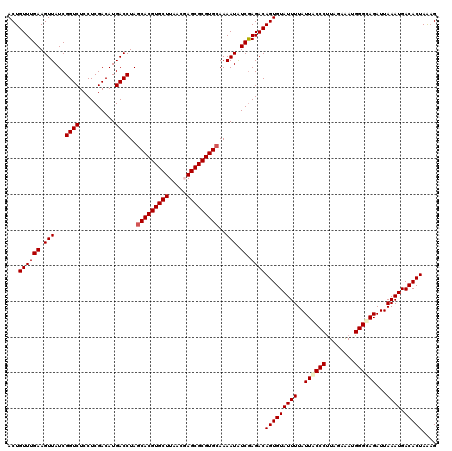
| Location | 20,422,390 – 20,422,510 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.72 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -31.95 |
| Energy contribution | -31.32 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.13 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20422390 120 + 20766785 AAUACACUGUCUCGACAUUUUGCACGCGCUCGUUAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCC .....(((((((((((((...(((((.(((.....))).)))))......))))))))....((..(((((......)))))..))......(((((.....)))))....))))).... ( -33.30) >DroPse_CAF1 35774 120 + 1 AAUACACUGUCUCGAUAUUUUGCACGCGCUCGUGAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCC .....(((((((((((((...(((((.(((.....))).)))))......))))))))....((..(((((......)))))..))......(((((.....)))))....))))).... ( -31.40) >DroGri_CAF1 39785 120 + 1 AAUACACUGUCUUGACAUUUUGCACGCGCUAGUUAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCC .....(((((((((((((((.(((((.(((.....))).)))))..(.((.(((((.(....)))))).((.(((....))).)))).)....))))))))))........))))).... ( -33.20) >DroWil_CAF1 44374 120 + 1 AAUACACUGUCUCGAUAUUUUGCACGCGCUGGUUAAGCACGUGUUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCC .....(((((((((((((...(((((.(((.....))).)))))......))))))))....((..(((((......)))))..))......(((((.....)))))....))))).... ( -31.00) >DroAna_CAF1 27797 120 + 1 AAUACACUGUCUCGAUAUUUUGCACGCGCUCGUGAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGUUGCAGGAACAAAUGGAUGUCAAGUUUAAGAAACAGUUUCC .....(((((.(((((.....(((((.(((.....))).)))))...))).(((((.(....))))))((((((..((.(((......))).))..))..))))....)).))))).... ( -30.30) >DroPer_CAF1 35548 120 + 1 AAUACACUGUCUCGAUAUUUUGCACGCGCUCGUGAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCC .....(((((((((((((...(((((.(((.....))).)))))......))))))))....((..(((((......)))))..))......(((((.....)))))....))))).... ( -31.40) >consensus AAUACACUGUCUCGAUAUUUUGCACGCGCUCGUGAAGCACGUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCC .....(((((((((((((...(((((.(((.....))).)))))......))))))))....((..(((((......)))))..))......(((((.....)))))....))))).... (-31.95 = -31.32 + -0.64)

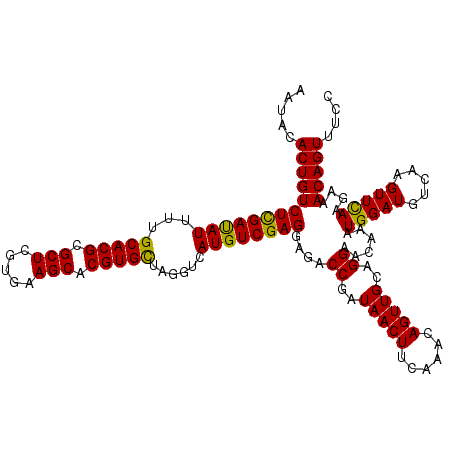
| Location | 20,422,390 – 20,422,510 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.72 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -35.83 |
| Energy contribution | -36.17 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.77 |
| SVM RNA-class probability | 0.999948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20422390 120 - 20766785 GGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUAACGAGCGCGUGCAAAAUGUCGAGACAGUGUAUU ....(((((......(((((..((..((.((((....)))).))..))))))).........((((((((......((((((((((...))))))))))...)))))))))))))..... ( -38.50) >DroPse_CAF1 35774 120 - 1 GGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUCACGAGCGCGUGCAAAAUAUCGAGACAGUGUAUU ....((((((((...(((((..((..((.((((....)))).))..)))))))...((((..........))))..((((((((((...))))))))))........))))))))..... ( -38.10) >DroGri_CAF1 39785 120 - 1 GGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUAACUAGCGCGUGCAAAAUGUCAAGACAGUGUAUU ....((((..((((((((((..((..((.((((....)))).))..))))))....((((..........))))..(((((((((.....)))))))))......))))))))))..... ( -35.70) >DroWil_CAF1 44374 120 - 1 GGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAACACGUGCUUAACCAGCGCGUGCAAAAUAUCGAGACAGUGUAUU ....((((((((...(((((..((..((.((((....)))).))..)))))))...((((..........))))...((((((((.....)))))))).........))))))))..... ( -33.10) >DroAna_CAF1 27797 120 - 1 GGAAACUGUUUCUUAAACUUGACAUCCAUUUGUUCCUGCAACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUCACGAGCGCGUGCAAAAUAUCGAGACAGUGUAUU ....((((((((...(((((..((..((.((((....)))).))..)))))))...((((..........))))..((((((((((...))))))))))........))))))))..... ( -38.00) >DroPer_CAF1 35548 120 - 1 GGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUCACGAGCGCGUGCAAAAUAUCGAGACAGUGUAUU ....((((((((...(((((..((..((.((((....)))).))..)))))))...((((..........))))..((((((((((...))))))))))........))))))))..... ( -38.10) >consensus GGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCACGUGCUUAACGAGCGCGUGCAAAAUAUCGAGACAGUGUAUU ....((((((((...(((((..((..((.((((....)))).))..)))))))...((((..........))))..(((((((((.....)))))))))........))))))))..... (-35.83 = -36.17 + 0.33)


| Location | 20,422,430 – 20,422,550 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.06 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -34.34 |
| Energy contribution | -34.15 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.67 |
| SVM RNA-class probability | 0.999936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20422430 120 + 20766785 GUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACGCGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUC ......((((.((((((.....((..(((((......)))))..))......(((((.....))))).((((((((((((((..(.....)..)))))))))))))).))))))..)))) ( -35.50) >DroVir_CAF1 59872 120 + 1 GUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACACGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUC ......((((.((((((.....((..(((((......)))))..))......(((((.....))))).((((((((((((((..(.....)..)))))))))))))).))))))..)))) ( -34.10) >DroWil_CAF1 44414 120 + 1 GUGUUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACGCGUAAAGAAUUGGAAAUUGUUUCAUCGACAAGGAUC ......((((.((((((.....((..(((((......)))))..))......(((((.....))))).((((((((((((((..(.....)..)))))))))))))).))))))..)))) ( -36.30) >DroMoj_CAF1 44064 120 + 1 GUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACACGAAAAGAAUUGGAAAUUGUUUCAUCAACACGGAUC ......((((.(((((.(....))))))..........((((..........(((((.....))))).((((((((((((((..(.....)..))))))))))))))..))))...)))) ( -32.40) >DroAna_CAF1 27837 120 + 1 GUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGUUGCAGGAACAAAUGGAUGUCAAGUUUAAGAAACAGUUUCCAACGCGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUC ......((((.((((((.....((..(((((......)))))..))......................((((((((((((((..(.....)..)))))))))))))).))))))..)))) ( -33.80) >DroPer_CAF1 35588 120 + 1 GUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACGCGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUC ......((((.((((((.....((..(((((......)))))..))......(((((.....))))).((((((((((((((..(.....)..)))))))))))))).))))))..)))) ( -35.50) >consensus GUGCUAGGUCAUGUCGAGGAGACCGAUAACUUCAAACAGUUGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACGCGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUC ......((((.((((((.....((..(((((......)))))..))......(((((.....))))).((((((((((((((..(.....)..)))))))))))))).))))))..)))) (-34.34 = -34.15 + -0.19)


| Location | 20,422,430 – 20,422,550 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.06 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -28.20 |
| Energy contribution | -28.07 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.995969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20422430 120 - 20766785 GAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGCGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCAC .....((.((((.((((((.((((((((.(.....).)))))))).))))))))))((((..((..((.((((....)))).))..))))))....((((..........))))..)).. ( -29.70) >DroVir_CAF1 59872 120 - 1 GAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGUGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCAC .....((.((((.((((((.((((((((.(.....).)))))))).))))))))))((((..((..((.((((....)))).))..))))))....((((..........))))..)).. ( -29.70) >DroWil_CAF1 44414 120 - 1 GAUCCUUGUCGAUGAAACAAUUUCCAAUUCUUUACGCGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAACAC ..((..((((((.((((((.((((((((.(.....).)))))))).))))))...(((((..((..((.((((....)))).))..)))))))..........)))))).))........ ( -29.10) >DroMoj_CAF1 44064 120 - 1 GAUCCGUGUUGAUGAAACAAUUUCCAAUUCUUUUCGUGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCAC .....((((((((((((((.((((((((.(.....).)))))))).))))))...(((((..((..((.((((....)))).))..))))))))))((((..........)))).))))) ( -30.80) >DroAna_CAF1 27837 120 - 1 GAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGCGUUGGAAACUGUUUCUUAAACUUGACAUCCAUUUGUUCCUGCAACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCAC .....((...((.((((((.((((((((.(.....).)))))))).))))))...(((((..((..((.((((....)))).))..))))))).))((((..........))))..)).. ( -29.30) >DroPer_CAF1 35588 120 - 1 GAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGCGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCAC .....((.((((.((((((.((((((((.(.....).)))))))).))))))))))((((..((..((.((((....)))).))..))))))....((((..........))))..)).. ( -29.70) >consensus GAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGCGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCAACUGUUUGAAGUUAUCGGUCUCCUCGACAUGACCUAGCAC .......(((((.((((((.((((((((.(.....).)))))))).))))))...(((((..((..((.((((....)))).))..)))))))..........)))))............ (-28.20 = -28.07 + -0.14)


| Location | 20,422,470 – 20,422,590 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.22 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -32.10 |
| Energy contribution | -31.77 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.998359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
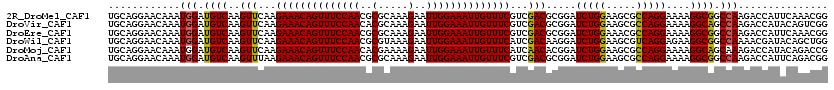
>2R_DroMel_CAF1 20422470 120 + 20766785 UGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACGCGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUCUGGAAGCGCCAGGAAAAGGCGGCCAAGACCAUUCAAACGG ............((((((((..(((...((((((((((((((..(.....)..))))))))))))))...))).......(((...((((.......)))).))).)).))))))..... ( -36.60) >DroVir_CAF1 59912 120 + 1 UGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACACGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUCUGGAAGCGCCAGGAAAAGGCAGCCAAGACCAUACAGUCGG ............(((.((((..(((...((((((((((((((..(.....)..))))))))))))))...))).....(((((.....)))))....)))).))).(((......))).. ( -35.20) >DroEre_CAF1 32535 120 + 1 UGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACGCGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUCUGGAAACGCCAGGAAAAGGCGGCCAAGACCAUUCAAACGG ............((((((((..(((...((((((((((((((..(.....)..))))))))))))))...))).......(((...((((.......)))).))).)).))))))..... ( -37.70) >DroWil_CAF1 44454 120 + 1 UGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACGCGUAAAGAAUUGGAAAUUGUUUCAUCGACAAGGAUCUGGAAGCGUCAGGAGAAGGCGGCCAAAACGAUACAGCUGG ..(((((.....(((((.....))))).((((((((((((((..(.....)..)))))))))))))).))..........(((..((.((....))..))..)))...........))). ( -30.20) >DroMoj_CAF1 44104 120 + 1 UGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACACGAAAAGAAUUGGAAAUUGUUUCAUCAACACGGAUCUGGAAGCGCCAGGAAAAGGCAGCAAAGACCAUACAGACCG ....((......(((((.....))))).((((((((((((((..(.....)..))))))))))))))........((.(((....(((((.......))).))..))))).......)). ( -31.40) >DroAna_CAF1 27877 120 + 1 UGCAGGAACAAAUGGAUGUCAAGUUUAAGAAACAGUUUCCAACGCGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUCUGGAAGCGCCAGGAAAAGGCGGCCAAGACCAUUCAGACGG ................((((..(((...((((((((((((((..(.....)..))))))))))))))...)))..((.(((((...((((.......)))).)).))))).....)))). ( -37.70) >consensus UGCAGGAACAAAUGGAUGUCAAGUUCAAGAAACAGUUUCCAACGCGCAAAGAAUUGGAAAUUGUUUCGUCGACGCGGAUCUGGAAGCGCCAGGAAAAGGCGGCCAAGACCAUACAGACGG ............(((.((((..(((...((((((((((((((..(.....)..))))))))))))))...))).....(((((.....)))))....)))).)))............... (-32.10 = -31.77 + -0.33)
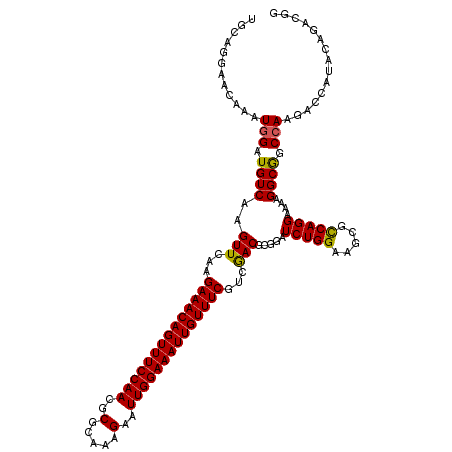
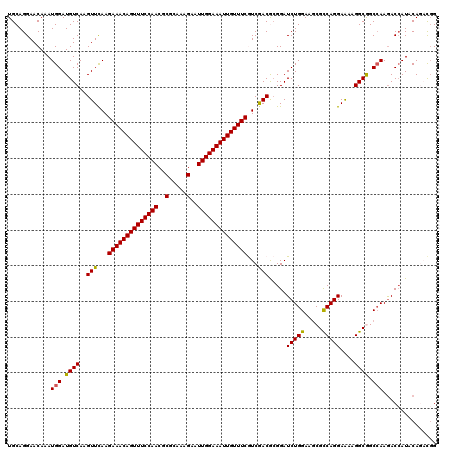
| Location | 20,422,470 – 20,422,590 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.22 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -26.91 |
| Energy contribution | -27.05 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
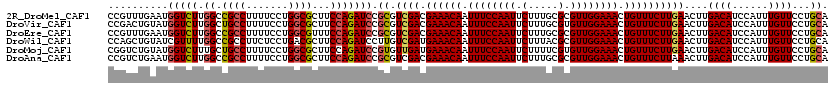
>2R_DroMel_CAF1 20422470 120 - 20766785 CCGUUUGAAUGGUCUUGGCCGCCUUUUCCUGGCGCUUCCAGAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGCGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCA ..((..((((((..((((.((((.......))))...))))..))...((((.((((((.((((((((.(.....).)))))))).))))))))))...............))))..)). ( -33.60) >DroVir_CAF1 59912 120 - 1 CCGACUGUAUGGUCUUGGCUGCCUUUUCCUGGCGCUUCCAGAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGUGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCA .((((.((..(((((.(((.(((.......))))))...))))).))))))..((((((.((((((((.(.....).)))))))).))))))..((((.((.....))...))))..... ( -34.60) >DroEre_CAF1 32535 120 - 1 CCGUUUGAAUGGUCUUGGCCGCCUUUUCCUGGCGUUUCCAGAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGCGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCA ..((..((((((..((((.((((.......))))...))))..))...((((.((((((.((((((((.(.....).)))))))).))))))))))...............))))..)). ( -33.60) >DroWil_CAF1 44454 120 - 1 CCAGCUGUAUCGUUUUGGCCGCCUUCUCCUGACGCUUCCAGAUCCUUGUCGAUGAAACAAUUUCCAAUUCUUUACGCGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCA ...((........(((((..((..((....)).))..)))))....((((((.((((((.((((((((.(.....).)))))))).))))))......)))))).............)). ( -25.90) >DroMoj_CAF1 44104 120 - 1 CGGUCUGUAUGGUCUUUGCUGCCUUUUCCUGGCGCUUCCAGAUCCGUGUUGAUGAAACAAUUUCCAAUUCUUUUCGUGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCA ..(((..((((((((..((.(((.......)))))....))).)))))..)))((((((.((((((((.(.....).)))))))).))))))..((((.((.....))...))))..... ( -32.50) >DroAna_CAF1 27877 120 - 1 CCGUCUGAAUGGUCUUGGCCGCCUUUUCCUGGCGCUUCCAGAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGCGUUGGAAACUGUUUCUUAAACUUGACAUCCAUUUGUUCCUGCA ..((..((((((..((((.((((.......))))...))))..))..(((((.((((((.((((((((.(.....).)))))))).))))))......)))))........))))..)). ( -33.00) >consensus CCGUCUGAAUGGUCUUGGCCGCCUUUUCCUGGCGCUUCCAGAUCCGCGUCGACGAAACAAUUUCCAAUUCUUUGCGCGUUGGAAACUGUUUCUUGAACUUGACAUCCAUUUGUUCCUGCA ..........(((((.((.((((.......))))...))))))).((.((((.((((((.((((((((.(.....).)))))))).))))))))))....((((......))))...)). (-26.91 = -27.05 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:13 2006