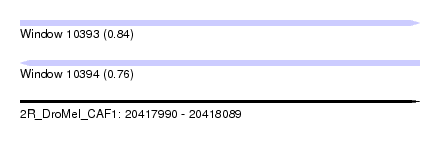
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,417,990 – 20,418,089 |
| Length | 99 |
| Max. P | 0.840739 |

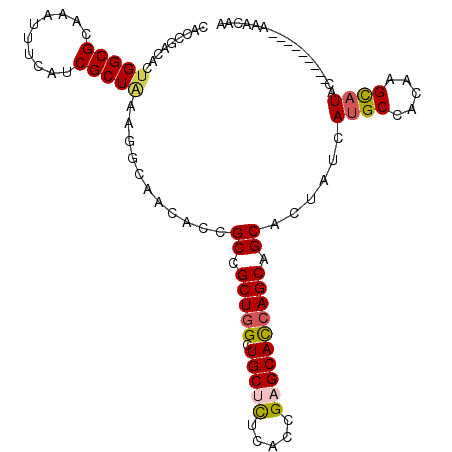
| Location | 20,417,990 – 20,418,089 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 83.53 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -21.04 |
| Energy contribution | -21.27 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20417990 99 + 20766785 CACCGACACUGGCGCAAAUUUCAUCGCUAAAAACCACCCCGCCGCUGGCUGCUCUUACCGAGCACCAGCAGCACUUUCAUGCCACAAGCAUUC---------AAGCAG .........((((((..........)).............((.(((((.(((((.....)))))))))).))........))))...((....---------..)).. ( -27.40) >DroVir_CAF1 37837 99 + 1 CACCGACACUGGCGCAAAUUUCAUCGCUAAAGGCAACACCGCCGCUGGCUGCCCUCACUGAGCACCAGCAGCACUAUCAUGCCACAAGCAUAC---------AAACCA ..........((.............(((...(((......)))(((((.(((.(.....).)))))))))))......((((.....))))..---------...)). ( -24.80) >DroGri_CAF1 34645 99 + 1 CACCAACACUGGCGCAAAUUUCAUCGCUAAAGGCAACACCGCCGCUGGCUGCCCUAACGGAGCAUCAGCAGCAUUAUCAUGCCACUAGCAUAC---------AGACCA ........(((..............(((...(((......)))(((((.(((((....)).)))))))))))......((((.....)))).)---------)).... ( -28.40) >DroWil_CAF1 39662 99 + 1 CACCAACACUGGCGCAAAUUUCAUCGCUAAAGGCCACACCGCCGCUGGCUGCUCUCACCGAGCAUCAGCAGCAUUUUCAUGCAACAAGUAUAC---------AGACAA ........(((..............(((...(((......)))(((((.(((((.....)))))))))))))......((((.....)))).)---------)).... ( -26.00) >DroMoj_CAF1 39327 102 + 1 CGCCGACACUGGCGCAAAUUUCAUCGCUGAAAGCAACACCGCCGCUGGCUGCUUUGACUGAGCACCAGCAGCACUAUCAUGCGACAAGCAUACAGC------AAACCA (((((....)))))...........((((...........((.(((((.(((((.....)))))))))).))......((((.....)))).))))------...... ( -34.50) >DroAna_CAF1 23525 108 + 1 CUCCGACACUGGCGCAAAUUUCAUCGCUCAAGGCCACCCCGCCGCUCGCUGCUCUUACGGAGCACCAGCAGCACUUCCAUGCCACCAGCCUCCAGCAGGCACAAGCAC .........(((((.................(((......)))(((.(((((((.....))))...)))))).......)))))...((((.....))))........ ( -27.00) >consensus CACCGACACUGGCGCAAAUUUCAUCGCUAAAGGCAACACCGCCGCUGGCUGCUCUCACCGAGCACCAGCAGCACUAUCAUGCCACAAGCAUAC_________AAACAA .........(((((..........)))))...........((.(((((.(((((.....)))))))))).))......((((.....))))................. (-21.04 = -21.27 + 0.23)

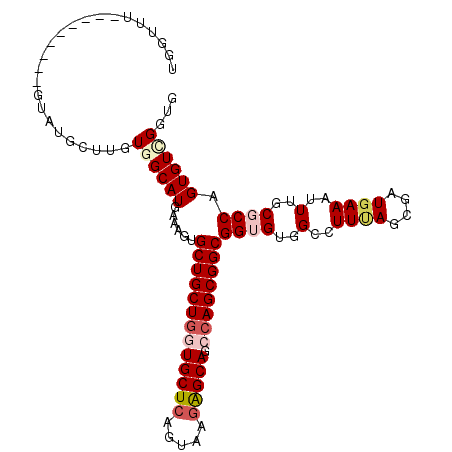
| Location | 20,417,990 – 20,418,089 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 83.53 |
| Mean single sequence MFE | -39.83 |
| Consensus MFE | -29.36 |
| Energy contribution | -30.45 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20417990 99 - 20766785 CUGCUU---------GAAUGCUUGUGGCAUGAAAGUGCUGCUGGUGCUCGGUAAGAGCAGCCAGCGGCGGGGUGGUUUUUAGCGAUGAAAUUUGCGCCAGUGUCGGUG ..((((---------..((((.....))))..))))(((((((((((((.....))))).))))))))((.(..(.(((((....))))).)..).)).......... ( -41.30) >DroVir_CAF1 37837 99 - 1 UGGUUU---------GUAUGCUUGUGGCAUGAUAGUGCUGCUGGUGCUCAGUGAGGGCAGCCAGCGGCGGUGUUGCCUUUAGCGAUGAAAUUUGCGCCAGUGUCGGUG ...((.---------.(.((((...((((..(((.((((((((((((((.....))))).))))))))).)))))))...)))))..)).....((((......)))) ( -40.90) >DroGri_CAF1 34645 99 - 1 UGGUCU---------GUAUGCUAGUGGCAUGAUAAUGCUGCUGAUGCUCCGUUAGGGCAGCCAGCGGCGGUGUUGCCUUUAGCGAUGAAAUUUGCGCCAGUGUUGGUG ......---------...((((((.((((..(((.((((((((.(((.((....)))))..)))))))).))))))).))))))..........((((......)))) ( -37.20) >DroWil_CAF1 39662 99 - 1 UUGUCU---------GUAUACUUGUUGCAUGAAAAUGCUGCUGAUGCUCGGUGAGAGCAGCCAGCGGCGGUGUGGCCUUUAGCGAUGAAAUUUGCGCCAGUGUUGGUG ......---------...((((....((((......(((((((.(((((.....)))))..)))))))((((..(..((((....))))..)..)))).)))).)))) ( -34.60) >DroMoj_CAF1 39327 102 - 1 UGGUUU------GCUGUAUGCUUGUCGCAUGAUAGUGCUGCUGGUGCUCAGUCAAAGCAGCCAGCGGCGGUGUUGCUUUCAGCGAUGAAAUUUGCGCCAGUGUCGGCG ......------((((.(((((.(.((((.((((.(((((((((((((.......)))).))))))))).))))..(((((....)))))..))))).))))))))). ( -41.00) >DroAna_CAF1 23525 108 - 1 GUGCUUGUGCCUGCUGGAGGCUGGUGGCAUGGAAGUGCUGCUGGUGCUCCGUAAGAGCAGCGAGCGGCGGGGUGGCCUUGAGCGAUGAAAUUUGCGCCAGUGUCGGAG ..((((((((((..(((((((..(..((((....))))..)..)).)))))..)).))).)))))((((((....)))...((((......))))))).......... ( -44.00) >consensus UGGUUU_________GUAUGCUUGUGGCAUGAAAGUGCUGCUGGUGCUCAGUAAGAGCAGCCAGCGGCGGUGUGGCCUUUAGCGAUGAAAUUUGCGCCAGUGUCGGUG ........................((((((......(((((((((((((.....))))).))))))))((((..(..((((....))))..)..)))).))))))... (-29.36 = -30.45 + 1.09)

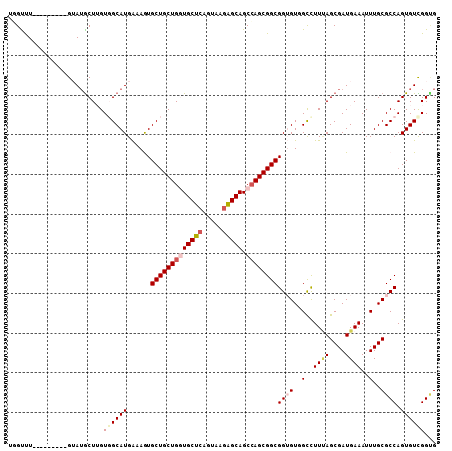
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:01 2006