| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,245,571 – 3,245,676 |
| Length | 105 |
| Max. P | 0.734939 |

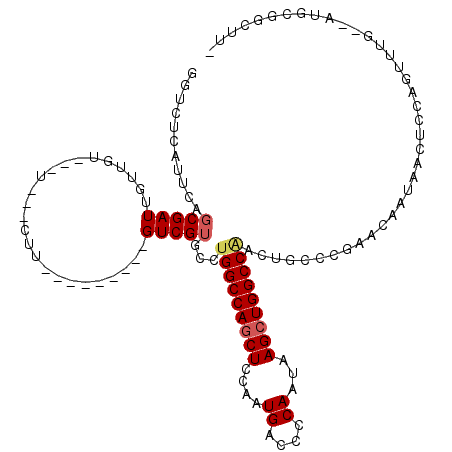
| Location | 3,245,571 – 3,245,676 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.90 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -15.36 |
| Energy contribution | -15.69 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.51 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3245571 105 + 20766785 GGUCUCAUUCAGCGAUUGUUGU---U---CUU---------GUCGUGCCUGGCCAACUCCAAUGACCCCAAUAAGCUGGCCAACUGCCCGAACAAUAACUCCAGUUUGUGGGGCGGCUUG .(((((((.....(((((..((---(---.((---------((((.((.((((((.((....((....))...)).))))))...)).)).)))).)))..))))).)))))))...... ( -29.60) >DroSec_CAF1 14715 94 + 1 GGUCACAUUCAGCGAUUGUUGU---U---UUU---------GUCGUGCCUGGCCAGCUCCAAUGACCCCAAUAAGCUGGCCAACUGGCCGAACAAUAGCUCC-----------CAGUUUG .............(((((..((---(---.((---------((((.((((((((((((....((....))...)))))))))...))))).)))).)))...-----------))))).. ( -29.70) >DroSim_CAF1 21511 94 + 1 CGUACCAUUCAGCGAUUGUUGU---U---CUU---------GUCGUGCCUGGCCAGCUCCAAUGACCCCAAUAAGCUGGCCAACUGCCCGAACAAUAGCUCC-----------CAGUUUG ...........(.(((((((((---(---(..---------((.((...(((((((((....((....))...))))))))))).))..))))))))).)).-----------)...... ( -26.60) >DroEre_CAF1 14371 102 + 1 GGUCUCAUUCAGCGAUUGUUGU---U---CUG---------GUCGUGCCUGGCCAGCUCCAAUGACCCCAAUAAGCUGGCCGACUACCCGAACAAUAACUCCAGCUUG--AUGCGGCCU- ((((.((((.((((((((((((---(---(.(---------((.((...(((((((((....((....))...))))))))))).))).))))))))).))..))).)--))).)))).- ( -42.50) >DroYak_CAF1 14669 111 + 1 GGACUCAUUCUGCGAUUGUUGU---U---CUUGUUGUUAUUGUCGUGACUGGCCAGCUCCGAUGACCCCAAUAAGCUGGCCGACUGCCCAAACAAUAACUCCAGUUUG--AUGCGGCUU- .(..(((..(((.(((((((((---(---...((.(((...((....)).((((((((....((....))...))))))))))).))...)))))))).)))))..))--)..).....- ( -30.00) >DroAna_CAF1 12328 98 + 1 UGUUCCAUUU--CGAUUAUUGCCGCUUUGGUU---------GUCGUGCUCGGCCAGCUCCAAUGACCCCAAUAAGCUGGCCA--------AACAAUAACUCCACUGUU--AUGCCAAAU- ((((......--((((....(((.....))).---------)))).....((((((((....((....))...)))))))).--------))))(((((......)))--)).......- ( -23.90) >consensus GGUCUCAUUCAGCGAUUGUUGU___U___CUU_________GUCGUGCCUGGCCAGCUCCAAUGACCCCAAUAAGCUGGCCAACUGCCCGAACAAUAACUCCAGUUUG__AUGCGGCUU_ ...........(((((.........................)))))...(((((((((....((....))...)))))))))...................................... (-15.36 = -15.69 + 0.33)

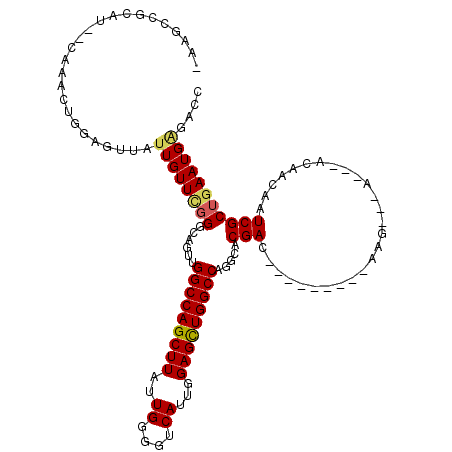
| Location | 3,245,571 – 3,245,676 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.90 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -19.96 |
| Energy contribution | -20.13 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 3245571 105 - 20766785 CAAGCCGCCCCACAAACUGGAGUUAUUGUUCGGGCAGUUGGCCAGCUUAUUGGGGUCAUUGGAGUUGGCCAGGCACGAC---------AAG---A---ACAACAAUCGCUGAAUGAGACC ...(((((((.((((((....))..))))..))))...((((((((((..((....))...))))))))))))).....---------...---.---...................... ( -31.90) >DroSec_CAF1 14715 94 - 1 CAAACUG-----------GGAGCUAUUGUUCGGCCAGUUGGCCAGCUUAUUGGGGUCAUUGGAGCUGGCCAGGCACGAC---------AAA---A---ACAACAAUCGCUGAAUGUGACC .......-----------..(((.(((((((((((...((((((((((..((....))...))))))))))))).))).---------...---.---...))))).))).......... ( -31.80) >DroSim_CAF1 21511 94 - 1 CAAACUG-----------GGAGCUAUUGUUCGGGCAGUUGGCCAGCUUAUUGGGGUCAUUGGAGCUGGCCAGGCACGAC---------AAG---A---ACAACAAUCGCUGAAUGGUACG ...((((-----------..(((.((((((((.((...((((((((((..((....))...)))))))))).)).))).---------...---.---...))))).)))...))))... ( -31.30) >DroEre_CAF1 14371 102 - 1 -AGGCCGCAU--CAAGCUGGAGUUAUUGUUCGGGUAGUCGGCCAGCUUAUUGGGGUCAUUGGAGCUGGCCAGGCACGAC---------CAG---A---ACAACAAUCGCUGAAUGAGACC -.((.(.(((--..(((..((....((((((.(((.((((((((((((..((....))...))))))))).)))...))---------).)---)---))))...)))))..))).).)) ( -37.90) >DroYak_CAF1 14669 111 - 1 -AAGCCGCAU--CAAACUGGAGUUAUUGUUUGGGCAGUCGGCCAGCUUAUUGGGGUCAUCGGAGCUGGCCAGUCACGACAAUAACAACAAG---A---ACAACAAUCGCAGAAUGAGUCC -.....((.(--((..(((..(((((((((..(((....(((((((((..((....))...))))))))).)))..))))))))).....(---(---.......)).)))..))))).. ( -32.30) >DroAna_CAF1 12328 98 - 1 -AUUUGGCAU--AACAGUGGAGUUAUUGUU--------UGGCCAGCUUAUUGGGGUCAUUGGAGCUGGCCGAGCACGAC---------AACCAAAGCGGCAAUAAUCG--AAAUGGAACA -..((.((..--....)).))(((..((((--------((((((((((..((....))...)))))))))))))).)))---------..(((...(((......)))--...))).... ( -31.20) >consensus _AAGCCGCAU__CAAACUGGAGUUAUUGUUCGGGCAGUUGGCCAGCUUAUUGGGGUCAUUGGAGCUGGCCAGGCACGAC_________AAG___A___ACAACAAUCGCUGAAUGAGACC .........................((((((((......(((((((((..((....))...))))))))).....(((...........................))))))))))).... (-19.96 = -20.13 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:46 2006