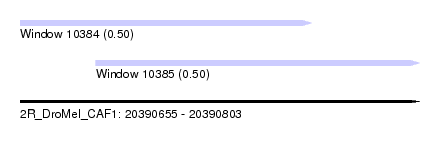
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,390,655 – 20,390,803 |
| Length | 148 |
| Max. P | 0.500000 |

| Location | 20,390,655 – 20,390,763 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.72 |
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -14.65 |
| Energy contribution | -14.82 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
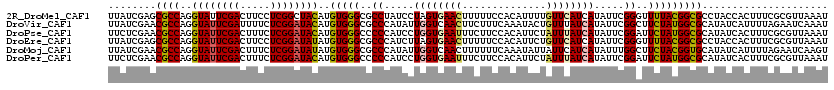
>2R_DroMel_CAF1 20390655 108 + 20766785 AUCUACAGCCGUUCCCCA------------UAGAUAACGAUUAUCGAGCGCCAGGUAUUCGACUUCCUCGGCUACAUGUGGGCGCCUAUCCUAGUGAACUUUUUCCACAUUUUGUUCAUC ..................------------..((((.((.....)).(((((..(((..(((.....)))..))).....))))).))))...((((((..............)))))). ( -20.94) >DroVir_CAF1 185 103 + 1 UUUACCAA-----------------AUCGACAGAUAACGAUUAUCGAACGCCAGGUAUUCGAUUUUCUCGGAUACAUGUGGGCGCCCAUAUUGGUCAACUUCUUUCAAAUACUGUUUAUC ........-----------------...(((((....((.....))...(((((((((((((.....)))))))).(((((....))))))))))................))))).... ( -23.60) >DroEre_CAF1 152 108 + 1 AUGUACAACUGCUGCCCC------------UAGAUAACGAUUAUCGAGCGCCAGGUAUUCGACUUCCUCGGAUAUAUGUGGGCGCCCAUCUUAGUGAACUUUUUCCACAUUCUGUUCAUC ..................------------..((((.....))))(.(((((..((((((((.....)))))))).....))))).)......((((((..............)))))). ( -23.44) >DroYak_CAF1 1 87 + 1 ---------------------------------AUAACGAUUAUCGAGCGUCAGGUAUUCGACUUCCUCGGAUACAUGUGGGCGCCCAUCCUAGUGAACUUUUUCCACAUUCUGUUCAUC ---------------------------------............(.(((((..((((((((.....)))))))).....))))).)......((((((..............)))))). ( -20.84) >DroMoj_CAF1 193 106 + 1 AUUGCCAA--------------ACAAUCGACAGAUAACGAUUAUCGAACGCCAGGUAUUCGACUUUCUCGGAUAUAUGUGGGCGCCCAUAUUGGUCAACUUUUUUCAAAUAUUAUUCAUC .(((((((--------------..(((((........)))))...(..((((..((((((((.....)))))))).....))))..)...)))).)))...................... ( -22.00) >DroPer_CAF1 125 120 + 1 AUCGAUUGCAUUUCCCCAUUUCACAAUGUAUAGAUAACGAUUCUCGAACGCCAGGUAUUCGACUUUCUCGGAUACAUGUGGGCCCCCAUCCUGGUGAAUUUCUUCCACAUUCUAUUUAUC (((...((((((............))))))..)))..((.....))..((((((((((((((.....))))))))..((((....)))).))))))........................ ( -26.40) >consensus AUCGACAA______________________UAGAUAACGAUUAUCGAACGCCAGGUAUUCGACUUCCUCGGAUACAUGUGGGCGCCCAUCCUAGUGAACUUUUUCCACAUUCUGUUCAUC .....................................((.....))..((((..((((((((.....)))))))).....)))).........((((((..............)))))). (-14.65 = -14.82 + 0.17)

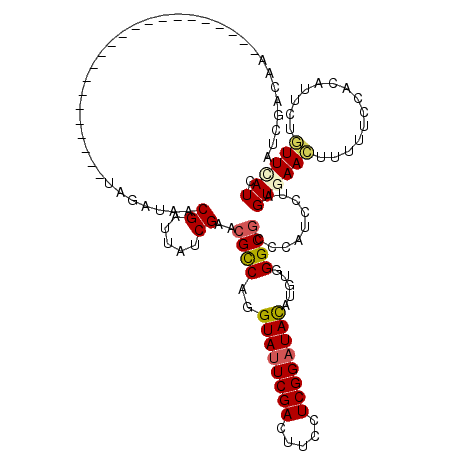
| Location | 20,390,683 – 20,390,803 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.83 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -22.60 |
| Energy contribution | -22.32 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 20390683 120 + 20766785 UUAUCGAGCGCCAGGUAUUCGACUUCCUCGGCUACAUGUGGGCGCCUAUCCUAGUGAACUUUUUCCACAUUUUGUUCAUCAUAUUCGGGUUUUACGGCGCCUACCACUUUCGCGUUAAAU ......(((((...(((..(((.....)))..)))..(((((((((.((((..((((((..............)))))).......)))).....))))))))).......))))).... ( -33.14) >DroVir_CAF1 208 120 + 1 UUAUCGAACGCCAGGUAUUCGAUUUUCUCGGAUACAUGUGGGCGCCCAUAUUGGUCAACUUCUUUCAAAUACUGUUUAUCAUAUUCGGCUUCUAUGGCGCAUAUCAUUUUAGAAUCAAAU ........((((..((((((((.....))))))))..(((((.(((.....((((.(((..............))).)))).....))).)))))))))..................... ( -26.74) >DroPse_CAF1 166 120 + 1 UUCUCGAACGCCAGGUAUUCGACUUUCUCGGAUACAUGUGGGCCCCCAUCCUGGUGAAUUUCUUCCACAUUCUAUUUAUCAUAUUCGGAUUCUAUGGCGCAUAUCACUUUCGCGUUAAAU ......(((((...((((((((.....))))))))(((((.(((...((((((((((((..............)))))))).....)))).....))).))))).......))))).... ( -29.94) >DroEre_CAF1 180 120 + 1 UUAUCGAGCGCCAGGUAUUCGACUUCCUCGGAUAUAUGUGGGCGCCCAUCUUAGUGAACUUUUUCCACAUUCUGUUCAUCAUAUUCGGGUUUUACGGCGCCUACCACUUUCGCGUUAAAU ......(((((...((((((((.....))))))))..(((((((((.......((((((..............))))))........(......)))))))))).......))))).... ( -33.74) >DroMoj_CAF1 219 120 + 1 UUAUCGAACGCCAGGUAUUCGACUUUCUCGGAUAUAUGUGGGCGCCCAUAUUGGUCAACUUUUUUCAAAUAUUAUUCAUCAUAUUUGGCUUCUACGGUGCAUAUCAUUUUAGAAUCAAGU .....((..(((((((((.(((.....)))(((..((((((....))))))..)))........................)))))))))(((((..(((.....)))..))))))).... ( -25.30) >DroPer_CAF1 165 120 + 1 UUCUCGAACGCCAGGUAUUCGACUUUCUCGGAUACAUGUGGGCCCCCAUCCUGGUGAAUUUCUUCCACAUUCUAUUUAUCAUAUUCGGAUUCUAUGGCGCAUAUCACUUUCGCGUUAAAU ......(((((...((((((((.....))))))))(((((.(((...((((((((((((..............)))))))).....)))).....))).))))).......))))).... ( -29.94) >consensus UUAUCGAACGCCAGGUAUUCGACUUUCUCGGAUACAUGUGGGCGCCCAUCCUGGUGAACUUCUUCCACAUUCUAUUCAUCAUAUUCGGAUUCUACGGCGCAUAUCACUUUCGCGUUAAAU ........((((..((((((((.....))))))))..(((((..((.....((((((((..............)))))))).....))..)))))))))..................... (-22.60 = -22.32 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:52 2006