
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,243,889 – 3,243,988 |
| Length | 99 |
| Max. P | 0.995750 |

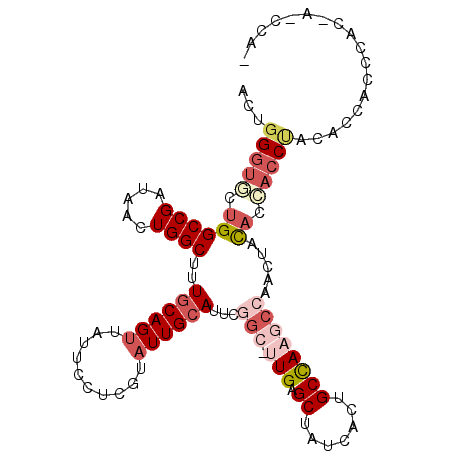
| Location | 3,243,889 – 3,243,988 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 76.46 |
| Mean single sequence MFE | -28.81 |
| Consensus MFE | -14.76 |
| Energy contribution | -16.65 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.51 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
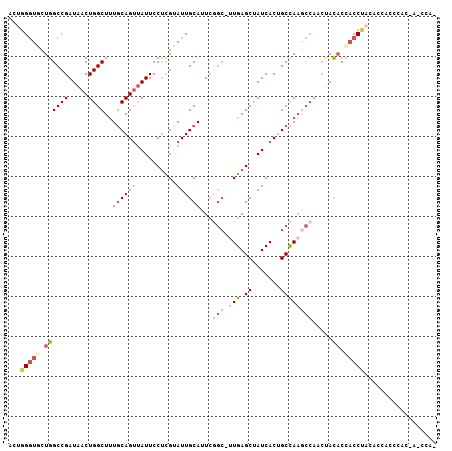
>2R_DroMel_CAF1 3243889 99 + 20766785 ACUGGGUGCUGGCCGAUAACUGGCUUUGCAGUUAUUCCUCGUAUUGCAUUCGGC-UUGAGCUAUCACUGCCAAGCCACCUACACCACCUACACCACCCAC------- ..((((((.((((((.....))))..((((((..........))))))...(((-(((.((.......))))))))..............)).)))))).------- ( -32.10) >DroSec_CAF1 13084 106 + 1 ACUUGGUGCUGGCCGAUAACUGGCUUUGCAGUUAUUCCUCGUAUUGCAUUCGGC-UUGAGCUAUCACUGCCAAGCCAACUACACCACCUACACCACCCACCAUCCAU ...(((((..((........((((((.(((((.((..((((....((.....))-.))))..)).))))).)))))).........))..)))))............ ( -30.03) >DroSim_CAF1 18098 106 + 1 ACUGGGUGCUGGCCGAUAACUGGCUUUGCAGUUAUUCCUCGUAUUGCAUUCGGC-UUGAGCUAUCACUGCCAAGCCAACUACACCACCUACACCACCAUCCAUCCAU ..((((((.(((..((((((((......))))))))...............(((-(((.((.......))))))))................))).))))))..... ( -30.90) >DroEre_CAF1 12779 93 + 1 ACUGGGUUCUGGCCGAUAACUGGCUUUGCAGUUAUUCCUUGUAUUGCAUUCAGC-UUGAGCUAUCACUGCCAAGCCACCCACACCACCC------------A-CCAA ..(((((...((........((((((.(((((.((..((((....((.....))-.))))..)).))))).))))))......))))))------------)-.... ( -24.74) >DroYak_CAF1 13096 100 + 1 CGUGGGUUCUGGCCGAUAACUGGCUUUGCAGUUAUUCCUCGAAUUGCAUUCGGG-CUGAGCUAUCACAGCCAUACCACCUACACAACCCAC-----CCACCA-CCAA .(((((((.((((((.....))))..(((((((........)))))))....((-(((........)))))..........)).)))))))-----......-.... ( -30.40) >DroAna_CAF1 10764 99 + 1 GCCGGCUUCUGGCCGAUAACUGGCUCUCCAGCCAUUCCCCGGAUUGCAAUGCACUAUGAGCAAUCACGGCUAACA-AAACAUUCCCCCUUCCCAAGCCUU------- ...(((((.((((((.....(((((....))))).......((((((............)))))).))))))...-.................)))))..------- ( -24.70) >consensus ACUGGGUGCUGGCCGAUAACUGGCUUUGCAGUUAUUCCUCGUAUUGCAUUCGGC_UUGAGCUAUCACUGCCAAGCCAACUACACCACCUACACCACCCAC_A_CCA_ ...(((((.((((((.....))))..((((((..........))))))...(((.(((.((.......)))))))).....)).))))).................. (-14.76 = -16.65 + 1.89)

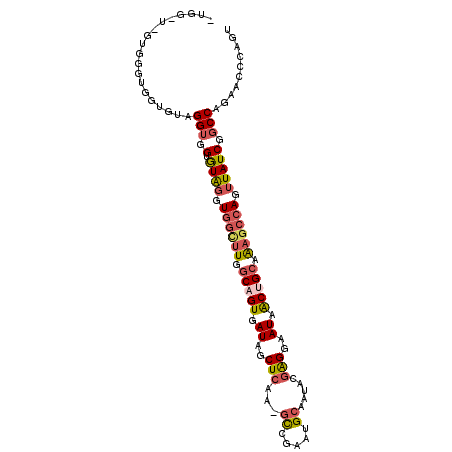
| Location | 3,243,889 – 3,243,988 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 76.46 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -19.36 |
| Energy contribution | -20.17 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
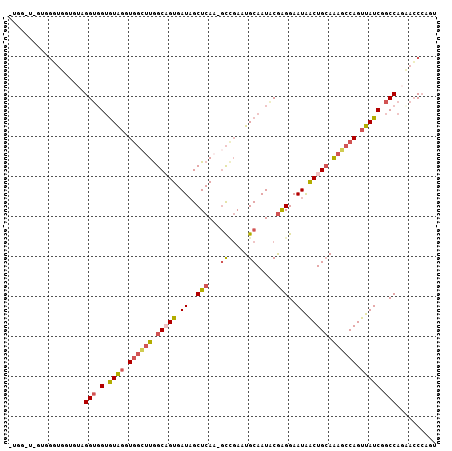
>2R_DroMel_CAF1 3243889 99 - 20766785 -------GUGGGUGGUGUAGGUGGUGUAGGUGGCUUGGCAGUGAUAGCUCAA-GCCGAAUGCAAUACGAGGAAUAACUGCAAAGCCAGUUAUCGGCCAGCACCCAGU -------.((((((.(....(((.((((..(((((((((.......)).)))-))))..)))).)))..((.(((((((......)))))))...))).)))))).. ( -38.50) >DroSec_CAF1 13084 106 - 1 AUGGAUGGUGGGUGGUGUAGGUGGUGUAGUUGGCUUGGCAGUGAUAGCUCAA-GCCGAAUGCAAUACGAGGAAUAACUGCAAAGCCAGUUAUCGGCCAGCACCAAGU .(((.((.(((.(((((..(((..(((((((((((((((.......)).)))-)))(....)............)))))))..)))...))))).))).)))))... ( -38.50) >DroSim_CAF1 18098 106 - 1 AUGGAUGGAUGGUGGUGUAGGUGGUGUAGUUGGCUUGGCAGUGAUAGCUCAA-GCCGAAUGCAAUACGAGGAAUAACUGCAAAGCCAGUUAUCGGCCAGCACCCAGU .....(((.((.((((....(((.((((.((((((((((.......)).)))-))))).)))).)))...((.((((((......)))))))).)))).)).))).. ( -37.30) >DroEre_CAF1 12779 93 - 1 UUGG-U------------GGGUGGUGUGGGUGGCUUGGCAGUGAUAGCUCAA-GCUGAAUGCAAUACAAGGAAUAACUGCAAAGCCAGUUAUCGGCCAGAACCCAGU ....-(------------(((((((((((.((((((.(((((..((((....-))))..((.....)).......))))).)))))).))))..)))...))))).. ( -30.80) >DroYak_CAF1 13096 100 - 1 UUGG-UGGUGG-----GUGGGUUGUGUAGGUGGUAUGGCUGUGAUAGCUCAG-CCCGAAUGCAAUUCGAGGAAUAACUGCAAAGCCAGUUAUCGGCCAGAACCCACG ....-..((((-----((.(((((.((((.((((..(((((.(....).)))-))....((((((((...))))...))))..)))).)))))))))...)))))). ( -36.90) >DroAna_CAF1 10764 99 - 1 -------AAGGCUUGGGAAGGGGGAAUGUUU-UGUUAGCCGUGAUUGCUCAUAGUGCAUUGCAAUCCGGGGAAUGGCUGGAGAGCCAGUUAUCGGCCAGAAGCCGGC -------..(((((.((.....(.(((((.(-(((.(((.......))).)))).))))).)...((((....(((((....)))))....))))))..)))))... ( -31.50) >consensus _UGG_U_GUGGGUGGUGUAGGUGGUGUAGGUGGCUUGGCAGUGAUAGCUCAA_GCCGAAUGCAAUACGAGGAAUAACUGCAAAGCCAGUUAUCGGCCAGAACCCAGU ...................(((.(.((((.((((((.(((((.((..(((...((.....)).....)))..)).))))).)))))).))))).))).......... (-19.36 = -20.17 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:44 2006