
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,373,510 – 20,373,615 |
| Length | 105 |
| Max. P | 0.920731 |

| Location | 20,373,510 – 20,373,615 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -16.85 |
| Energy contribution | -15.97 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.920731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
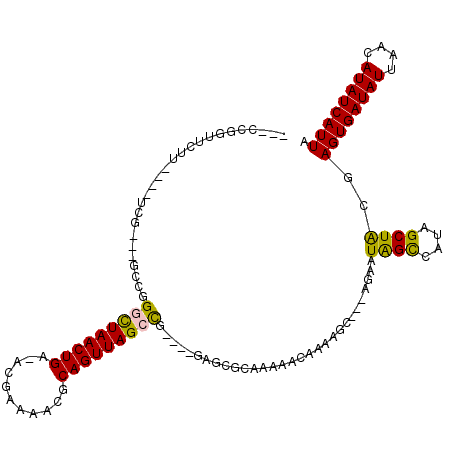
>2R_DroMel_CAF1 20373510 105 + 20766785 AACCCGGUUCUU----UCG---GCCCGGCUAACUGA-ACUAAACCGCAGUUAGGCA----GAGCGCAAAAACAAAGGC---AGAAUGGCCAUAGCUGCGAGUGAUAUUAACAUAUCAUUA ...((((..(..----..)---..))))........-.......(((((((((((.----...(((..........))---.)....))).))))))))((((((((....)))))))). ( -29.70) >DroVir_CAF1 122250 108 + 1 ---UUUUUUUUUGC--CUG---GCCGGGUUAACUGAAACGAAAACGCAGUUAGCCC----GAGCGCAAAAACAAAAGCAGCAGCAUAGUUAUAGCUACGAGUGAUAUUAACAUAUCAUUA ---(((.(((((((--...---(((((((((((((...........))))))))))----).))))))))).))).((....)).((((....))))..((((((((....)))))))). ( -32.10) >DroGri_CAF1 114600 106 + 1 ---ACAGUGUUACCUUUUC---GCCGGUUUAACUGAAACGAAAACGCAGUUAGUU-----GAGCGCAAAAACAAAAGC---AGCAUAGUUAUAGCUACGAGUGAUAUUAACAUAUCAUUA ---...((((((.....((---((((((((((((((..((....))...))))))-----))))..............---....((((....)))))).))))...))))))....... ( -21.60) >DroEre_CAF1 97542 105 + 1 AACCCGGUUCUU----UCG---GCCCGGCUAACUGA-ACUAAACCGCAGUUAGCCG----GAGCGCAAAAACAAAGGC---AGAAUGGCCAUAGCUGCGAGUGAUAUUAACAUAUCAUUA ..(.(((((...----...---(((((((((((((.-.........))))))))))----).))...........(((---......)))..))))).)((((((((....)))))))). ( -35.10) >DroMoj_CAF1 135709 108 + 1 ---UCAGUUUUUGC--UUG---GCCGGGUUAACUGAAACGAAAACGCAGUUAGCUG----GAGCGCAAAAACAAAAGCAACAGCAUAGUUAUAGCUACGAGUGAUAUUAACAUAUCAUUA ---...((((((((--...---(((.(((((((((...........))))))))).----).))))))))))....((....)).((((....))))..((((((((....)))))))). ( -29.90) >DroAna_CAF1 89592 109 + 1 ---GCGGUCCGU----CCGUAACCCAGGCUAACUGA-ACUAAAACGCAGUUAGCCGACUUGAGCGCAAAAACAAAAGC---AGAAUGGCCAUAGCUGCGAGUGAUAUUAACAUAUCAUUA ---(((.((.((----(.(.....).(((((((((.-.........))))))))))))..)).)))..........((---((...........)))).((((((((....)))))))). ( -30.60) >consensus ___CCGGUUCUU____UCG___GCCGGGCUAACUGA_ACGAAAACGCAGUUAGCCG____GAGCGCAAAAACAAAAGC___AGAAUAGCCAUAGCUACGAGUGAUAUUAACAUAUCAUUA ..........................(((((((((...........)))))))))..............................((((....))))..((((((((....)))))))). (-16.85 = -15.97 + -0.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:45 2006