
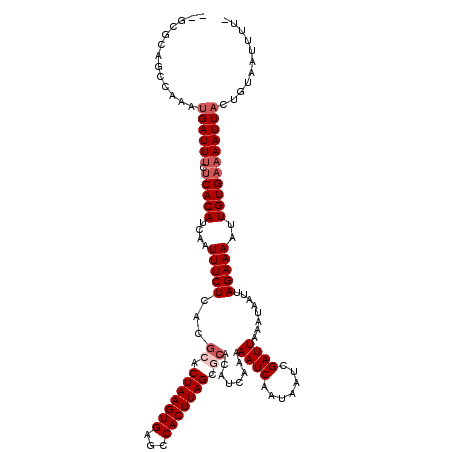
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,366,805 – 20,366,923 |
| Length | 118 |
| Max. P | 0.847349 |

| Location | 20,366,805 – 20,366,923 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.09 |
| Mean single sequence MFE | -19.55 |
| Consensus MFE | -16.40 |
| Energy contribution | -17.40 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
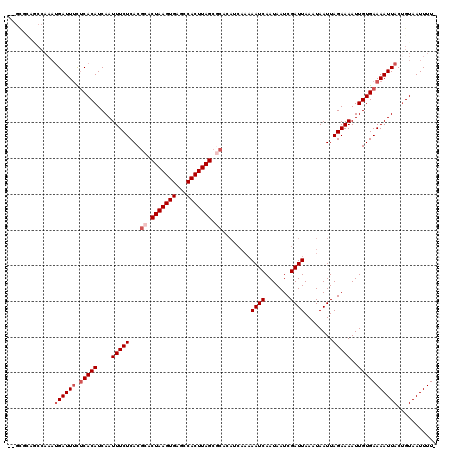
>2R_DroMel_CAF1 20366805 118 + 20766785 U-GCGCAGCCAAAUGAUUCUGCACAUCAAUUUCUCACGCACUAAGUGAGCCACUUAGCACACAUCAAAAAUCAAUAAUCGAUUAAAUAAUUAGAAAAUUGUGAAAAUUACUGUAAUUUU- .-..((((..........))))(((..(((((.(((((..(((((((...)))))))..)........((((.......))))................)))))))))..)))......- ( -17.10) >DroVir_CAF1 112082 117 + 1 --GCAGAGCCAAAUGAUUUCUCACAUCAAUUUCUCACGCACUAAGUGAGCCACUUAGCGCACAUCAAAAAUCAAUAAUCGAUUAAAUAAUUAGAAAAUUGUGAAAAUUACUGUAAUUUU- --((((.......((((((.(((((....(((((...((.(((((((...))))))).))........((((.......))))........)))))..)))))))))))))))......- ( -22.01) >DroGri_CAF1 106256 120 + 1 GCGCACAUCCAAAUGAUUUCUCACAUCAAUUUCUCACGCACUAAGUGAGCCACUUAGCGCACAUCAAAAAUCAAUAAUCGAUUAAAUAAUUAGAAAAUUGUGAAAAUUACAGUAAUUUUG .............((((((.(((((....(((((...((.(((((((...))))))).))........((((.......))))........)))))..)))))))))))........... ( -20.50) >DroWil_CAF1 159206 107 + 1 A-UUGUGGCCAAAUGAUUUCUCACAUCAAUUUCUCACGCACUAAGUGAGCCACUUAG-----------AAUCAAUAAUCGAUUAAAUAAUUAGAAAAUUGUGAAAAUUACUGUAAUUUU- (-(((..(.....((((((.(((((....(((((......(((((((...)))))))-----------((((.......))))........)))))..))))))))))))..))))...- ( -17.30) >DroMoj_CAF1 126186 105 + 1 --------------GAUUUCUCACAUCAAUUUCUCACGCACUAAGUGAGCCACUUAGCGCACAUCAAAAAUCAAUAAUCGAUUAAAUAAUUAGAAAAUUGUGAAAAUUACUGUAAUUUU- --------------(((((.(((((....(((((...((.(((((((...))))))).))........((((.......))))........)))))..))))))))))...........- ( -19.40) >DroPer_CAF1 97792 118 + 1 G-GCGCAACCAAAUGAUUUCUCACAUCAAUUUCUCACGCACUAAGUGAGCCACUUAGCGCACAUCAAAAAUCAAUAAUCGAUUAAAUAAUUAGAAAAUUGUGAAAAUUACUGUAAUUUU- (-(.....))...((((((.(((((....(((((...((.(((((((...))))))).))........((((.......))))........)))))..)))))))))))..........- ( -21.00) >consensus __GCGCAGCCAAAUGAUUUCUCACAUCAAUUUCUCACGCACUAAGUGAGCCACUUAGCGCACAUCAAAAAUCAAUAAUCGAUUAAAUAAUUAGAAAAUUGUGAAAAUUACUGUAAUUUU_ .............((((((.(((((....(((((...((.(((((((...))))))).))........((((.......))))........)))))..)))))))))))........... (-16.40 = -17.40 + 1.00)

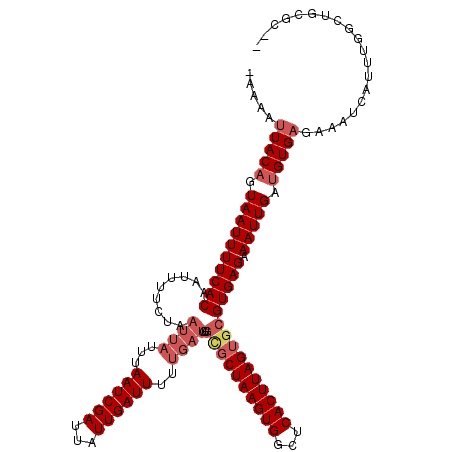
| Location | 20,366,805 – 20,366,923 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.09 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -23.06 |
| Energy contribution | -24.45 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
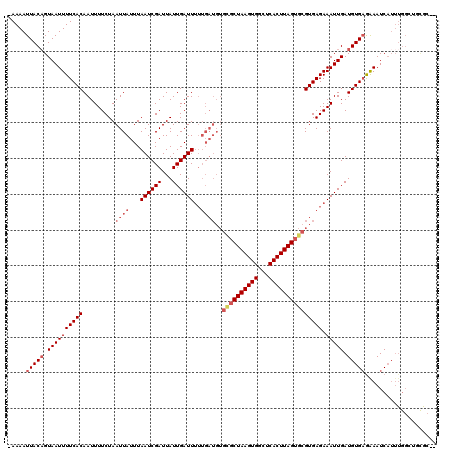
>2R_DroMel_CAF1 20366805 118 - 20766785 -AAAAUUACAGUAAUUUUCACAAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGUGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGCAGAAUCAUUUGGCUGCGC-A -.......((((..(((((((.........((((...((((((...))))))..))))..(..(((((((...)))))))..)))))))))))).(((((((..........))))))-) ( -31.10) >DroVir_CAF1 112082 117 - 1 -AAAAUUACAGUAAUUUUCACAAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAGAAAUCAUUUGGCUCUGC-- -.......(((...(((((((.........((((...((((((...))))))..))))..((((((((((...))))))))))))))))).(..(.((((....)))))..)..))).-- ( -28.60) >DroGri_CAF1 106256 120 - 1 CAAAAUUACUGUAAUUUUCACAAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAGAAAUCAUUUGGAUGUGCGC ..........(((.(((((((.........((((...((((((...))))))..))))..((((((((((...)))))))))))))))))(((.(.((((....)))).).))).))).. ( -30.10) >DroWil_CAF1 159206 107 - 1 -AAAAUUACAGUAAUUUUCACAAUUUUCUAAUUAUUUAAUCGAUUAUUGAUU-----------CUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAGAAAUCAUUUGGCCACAA-U -.......((((..(((((((................((((((...))))))-----------(((((((...)))))))...)))))))))))..((((....))))(((....)))-. ( -18.50) >DroMoj_CAF1 126186 105 - 1 -AAAAUUACAGUAAUUUUCACAAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAGAAAUC-------------- -....(((((.((((((((((.........((((...((((((...))))))..))))..((((((((((...))))))))))))))).))))).)))))......-------------- ( -28.10) >DroPer_CAF1 97792 118 - 1 -AAAAUUACAGUAAUUUUCACAAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAGAAAUCAUUUGGUUGCGC-C -.........(((.(((((((.........((((...((((((...))))))..))))..((((((((((...)))))))))))))))))((..(.((((....)))))..)))))..-. ( -29.70) >consensus _AAAAUUACAGUAAUUUUCACAAUUUUCUAAUUAUUUAAUCGAUUAUUGAUUUUUGAUGUGCGCUAAGUGGCUCACUUAGUGCGUGAGAAAUUGAUGUGAGAAAUCAUUUGGCUGCGC__ .....(((((.((((((((((.........((((...((((((...))))))..))))..((((((((((...))))))))))))))).))))).))))).................... (-23.06 = -24.45 + 1.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:41 2006