
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,362,499 – 20,362,592 |
| Length | 93 |
| Max. P | 0.637821 |

| Location | 20,362,499 – 20,362,592 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.18 |
| Mean single sequence MFE | -15.97 |
| Consensus MFE | -11.21 |
| Energy contribution | -11.71 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.637821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
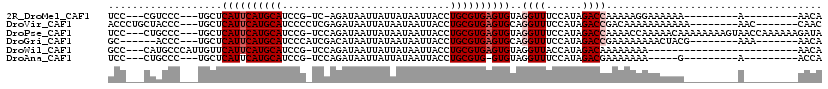
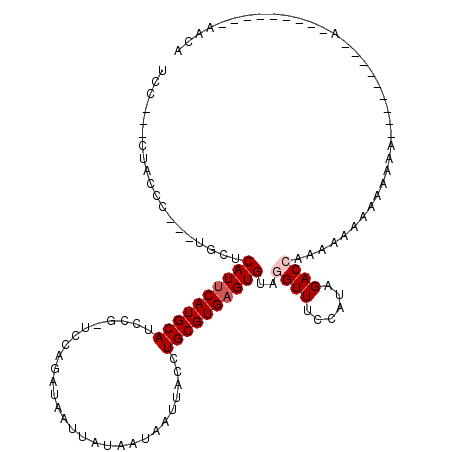
>2R_DroMel_CAF1 20362499 93 + 20766785 UCC---CGUCCC---UGCUCAUUCAUGCAUCCG-UC-AGAUAAUUAUUAUAAUUACCUGCGUGAGUGUAGGUUUCCAUAGACCAAAAAGGAAAAAA---------A---------AACA ...---....((---(...((((((((((((..-..-.))((((((...))))))..))))))))))..((((......))))....)))......---------.---------.... ( -17.60) >DroVir_CAF1 107423 101 + 1 ACCCUGCUACCC---UGCUCAUUCAUGCAUCCCCUCGAGAUAAUUAUAAUAAUUACCUGCGUGAGUGCAGGUUUCCAUAGACCGACAAAAAAAAAAA--------AAC-------CAAC ..........((---(((..(((((((((((.....))..((((((...))))))..))))))))))))))..........................--------...-------.... ( -16.80) >DroPse_CAF1 93088 112 + 1 UCC---CUGCCC---UGCUCAUUCAUGCAUCCG-UCCAGAUAAUUAUAAUAAUUACCUGCGUGAGUGUAGGUUUCCAUAGACCAAAACCAAAAACAAAAAAAAGUAACCAAAAAAGAUA ...---......---((((((((((((((((..-....))((((((...))))))..))))))))))..((((......))))...................))))............. ( -16.40) >DroGri_CAF1 102264 95 + 1 GC------ACCC---UGCUCAUUCAUGCAUCCCAUCGACAUAAUUAUAAUAAUUACCUGCGUGAGUGCAGGUUUCCAUAGACCGAAAAAAAACUACG--------AAA-------AACA ..------..((---(((..(((((((((((.....))..((((((...))))))..))))))))))))))..........................--------...-------.... ( -16.50) >DroWil_CAF1 150599 90 + 1 GCC---CAUGCCCAUUGUUCAUUCAUGCAUCCG-UCCAGAUAAUUAUUAUAAUUACCUGCGUGAGUGUAGGUUACCAUAGACAAAAAAAA-------------------------AACA ...---........(((((((((((((((((..-....))((((((...))))))..))))))))))..((...))...)))))......-------------------------.... ( -13.10) >DroAna_CAF1 77643 88 + 1 UCC---CUGCCC---UGCUCAUUCAUGCAUCCG-UCCAGAUAAUUAUUAUAAUUACCUGCGUG-GUGUAGGUUUCCAUAGACGAAAAAAA-----G---------A---------ACCA ..(---((((..---(((........))).(((-..(((.((((((...)))))).)))..))-).)))))...................-----.---------.---------.... ( -15.40) >consensus UCC___CUACCC___UGCUCAUUCAUGCAUCCG_UCCAGAUAAUUAUAAUAAUUACCUGCGUGAGUGUAGGUUUCCAUAGACCAAAAAAAAAAAAA_________A_________AACA ...................((((((((((............................))))))))))..((((......)))).................................... (-11.21 = -11.71 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:38 2006