
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 20,353,476 – 20,353,577 |
| Length | 101 |
| Max. P | 0.789600 |

| Location | 20,353,476 – 20,353,577 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.43 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -13.30 |
| Energy contribution | -14.22 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
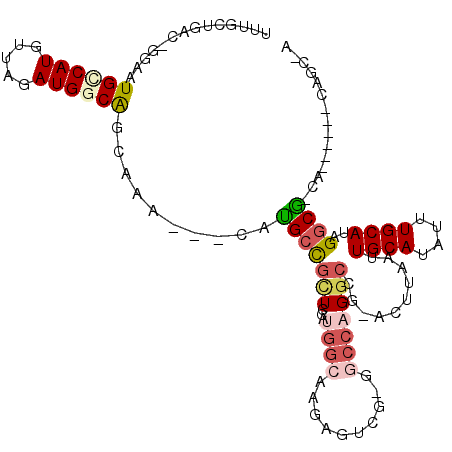
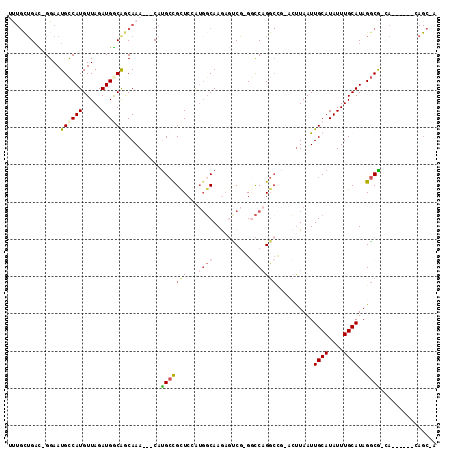
>2R_DroMel_CAF1 20353476 101 + 20766785 UUUGCUGAC-GGAAUGCCAUGUUAGAUGGCAGCAAA---CAUGCCGUUCGAUGGCAAGAGUCG-GGCCAGGCCG-ACUUAAUUGCAUAUUUGCAUAGGCG-CA------GAGC-A (((((....-....((((((....(((((((.....---..)))))))..)))))).((((((-(......)))-))))...((((....)))).....)-))------))..-. ( -34.10) >DroPse_CAF1 82553 102 + 1 UUUGCUGAU-GGAAUGCCAUGUUAGAUGACGGCAAA---CAUGCCGCUCCAUGGCAAGAGUCGGGGCCAGGCUG-ACUUAAUUGCAUAUUUGCAUAGGCG-CA------CAGC-A ..(((((((-(.(((((((((...((.(.(((((..---..))))))))))))))..(((((((.......)))-)))).))).)))...(((......)-))------))))-) ( -35.10) >DroWil_CAF1 134481 105 + 1 ACUGUUGACUGGAAUGCCAUGUUAGAUGGCAACAAACAACAUGCCGCUCAGGCUCAAACUCCG-GCUCAGGCCAAAGUUAAUUGCAUAUUUGCAUAGGCAAGA------GAG--- ..(((((..((...((((((.....)))))).))..)))))((((((....))...((((..(-((....)))..))))...((((....))))..))))...------...--- ( -29.90) >DroMoj_CAF1 110049 90 + 1 UC---AGCU-GGAAUGUCAUGUUAGAUGGCAACAAA---CAUGACGCUCCAUGGCAAAAAGCU-GACCAG-------CUAAUUGCAUAUUUGCAUAGCCA-GC------C---GA ((---.(((-(((.(((((((((...((....))))---))))))).))))((((....((((-....))-------))...((((....))))..))))-))------.---)) ( -33.40) >DroAna_CAF1 69203 108 + 1 CUUGCUGAC-GGAAUGCCAUGUUAGAUGGCAGCAAA---CACGCUGCUCGAUGGCAAGAGCCGGGGCCCGGCCG-ACUUAAUUGCAUAUUUGCAUAGGCG-CACCAGAAAAGC-A ..((((...-((..((((.(((((...((((((...---...))))))...))))).(.(((((...)))))).-.......((((....))))..))))-..)).....)))-) ( -35.90) >DroPer_CAF1 82610 102 + 1 UUUGCUGAU-GGAAUGCCAUGUUAGAUGACGGCAAA---CAUGCCGCUCCAUGGCAAGAGUCGGGGCCAGGCUG-ACUUAAUUGCAUAUUUGCAUAGGCG-CA------CAGC-A ..(((((((-(.(((((((((...((.(.(((((..---..))))))))))))))..(((((((.......)))-)))).))).)))...(((......)-))------))))-) ( -35.10) >consensus UUUGCUGAC_GGAAUGCCAUGUUAGAUGGCAGCAAA___CAUGCCGCUCCAUGGCAAGAGUCG_GGCCAGGCCG_ACUUAAUUGCAUAUUUGCAUAGGCG_CA______CAGC_A ..............((((((.....))))))..........(((((((...((((..........)))))))..........((((....))))..))))............... (-13.30 = -14.22 + 0.92)

| Location | 20,353,476 – 20,353,577 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.43 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -13.78 |
| Energy contribution | -13.90 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
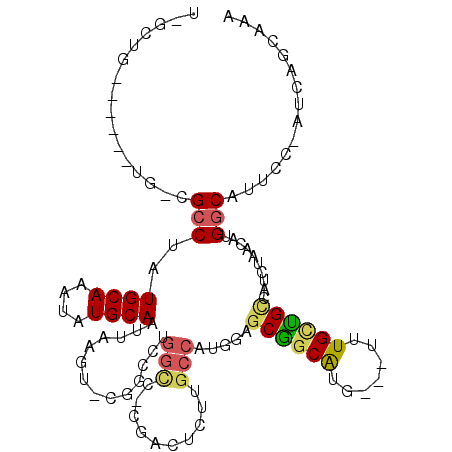
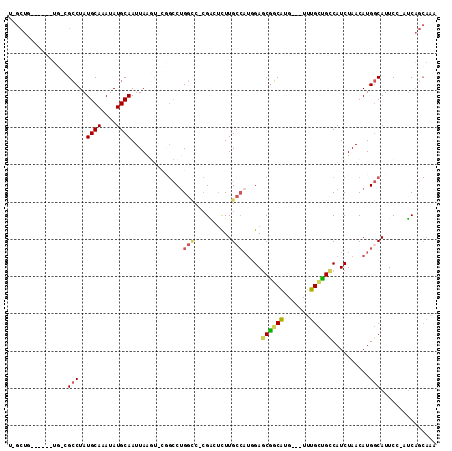
>2R_DroMel_CAF1 20353476 101 - 20766785 U-GCUC------UG-CGCCUAUGCAAAUAUGCAAUUAAGU-CGGCCUGGCC-CGACUCUUGCCAUCGAACGGCAUG---UUUGCUGCCAUCUAACAUGGCAUUCC-GUCAGCAAA (-(((.------.(-((.....((((((((((.......(-(((..((((.-........))))))))...)))))---)))))((((((.....))))))...)-)).)))).. ( -32.10) >DroPse_CAF1 82553 102 - 1 U-GCUG------UG-CGCCUAUGCAAAUAUGCAAUUAAGU-CAGCCUGGCCCCGACUCUUGCCAUGGAGCGGCAUG---UUUGCCGUCAUCUAACAUGGCAUUCC-AUCAGCAAA (-((((------((-.......((((((((((......((-(.....)))..((.((((......)))))))))))---))))).(((((.....)))))....)-).))))).. ( -31.30) >DroWil_CAF1 134481 105 - 1 ---CUC------UCUUGCCUAUGCAAAUAUGCAAUUAACUUUGGCCUGAGC-CGGAGUUUGAGCCUGAGCGGCAUGUUGUUUGUUGCCAUCUAACAUGGCAUUCCAGUCAACAGU ---...------...((((..((((....))))...(((((((((....))-)))))))...((....))))))(((((.(((.((((((.....))))))...))).))))).. ( -32.80) >DroMoj_CAF1 110049 90 - 1 UC---G------GC-UGGCUAUGCAAAUAUGCAAUUAG-------CUGGUC-AGCUUUUUGCCAUGGAGCGUCAUG---UUUGUUGCCAUCUAACAUGACAUUCC-AGCU---GA ((---(------((-(((....((...(((((((..((-------((....-))))..))).))))..))((((((---((.(.......).))))))))...))-))))---)) ( -34.10) >DroAna_CAF1 69203 108 - 1 U-GCUUUUCUGGUG-CGCCUAUGCAAAUAUGCAAUUAAGU-CGGCCGGGCCCCGGCUCUUGCCAUCGAGCAGCGUG---UUUGCUGCCAUCUAACAUGGCAUUCC-GUCAGCAAG (-(((....(((..-.......((((((((((........-.((((((...))))))..(((......))))))))---)))))((((((.....))))))..))-)..)))).. ( -35.70) >DroPer_CAF1 82610 102 - 1 U-GCUG------UG-CGCCUAUGCAAAUAUGCAAUUAAGU-CAGCCUGGCCCCGACUCUUGCCAUGGAGCGGCAUG---UUUGCCGUCAUCUAACAUGGCAUUCC-AUCAGCAAA (-((((------((-.......((((((((((......((-(.....)))..((.((((......)))))))))))---))))).(((((.....)))))....)-).))))).. ( -31.30) >consensus U_GCUG______UG_CGCCUAUGCAAAUAUGCAAUUAAGU_CGGCCUGGCC_CGACUCUUGCCAUGGAGCGGCAUG___UUUGCUGCCAUCUAACAUGGCAUUCC_AUCAGCAAA ................(((..((((....))))..............(((..........))).....((((((.......))))))..........)))............... (-13.78 = -13.90 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:12:37 2006